Microbiome profile of the amniotic fluid as a predictive biomarker of perinatal outcome
- PMID: 28939908
- PMCID: PMC5610236
- DOI: 10.1038/s41598-017-11699-8
Microbiome profile of the amniotic fluid as a predictive biomarker of perinatal outcome
Abstract
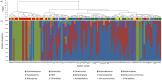
Chorioamnionitis (CAM), an inflammation of the foetal membranes due to infection, is associated with preterm birth and poor perinatal prognosis. The present study aimed to determine whether CAM can be diagnosed prior to delivery based on the bacterial composition of the amniotic fluid (AF). AF samples from 79 patients were classified according to placental inflammation: Stage III (n = 32), CAM; Stage II (n = 27), chorionitis; Stage 0-I (n = 20), sub-chorionitis or no neutrophil infiltration; and normal AF in early pregnancy (n = 18). Absolute quantification and sequencing of 16S rDNA showed that in Stage III, the 16S rDNA copy number was significantly higher and the α-diversity index lower than those in the other groups. In principal coordinate analysis, Stage III formed a separate cluster from Stage 0-I, normal AF, and blank. Forty samples were classified as positive for microbiomic CAM (miCAM) defined by the presence of 11 bacterial species that were found to be significantly associated with CAM and some parameters of perinatal prognosis. The diagnostic accuracy for CAM according to miCAM was: sensitivity, approximately 94%, and specificity, 79-87%. Our findings indicate the possibility of predicting CAM prior to delivery based on the AF microbiome profile.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

