Influence of OATPs on Hepatic Disposition of Erlotinib Measured With Positron Emission Tomography
- PMID: 28940241
- PMCID: PMC6083370
- DOI: 10.1002/cpt.888
Influence of OATPs on Hepatic Disposition of Erlotinib Measured With Positron Emission Tomography
Abstract
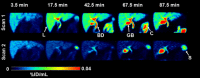
To assess the hepatic disposition of erlotinib, we performed positron emission tomography (PET) scans with [11 C]erlotinib in healthy volunteers without and with oral pretreatment with a therapeutic erlotinib dose (300 mg). Erlotinib pretreatment significantly decreased the liver exposure to [11 C]erlotinib with a concomitant increase in blood exposure, pointing to the involvement of a carrier-mediated hepatic uptake mechanism. Using cell lines overexpressing human organic anion-transporting polypeptides (OATPs) 1B1, 1B3, or 2B1, we show that [11 C]erlotinib is selectively transported by OATP2B1. Our data suggest that at PET microdoses hepatic uptake of [11 C]erlotinib is mediated by OATP2B1, whereas at therapeutic doses OATP2B1 transport is saturated and hepatic uptake occurs mainly by passive diffusion. We propose that [11 C]erlotinib may be used as a hepatic OATP2B1 probe substrate and erlotinib as an OATP2B1 inhibitor in clinical drug-drug interaction studies, allowing the contribution of OATP2B1 to the hepatic uptake of drugs to be revealed.
© 2017 The Authors Clinical Pharmacology & Therapeutics published by Wiley Periodicals, Inc. on behalf of American Society for Clinical Pharmacology and Therapeutics.
Figures




Similar articles
-
Effect of Rifampicin on the Distribution of [11C]Erlotinib to the Liver, a Translational PET Study in Humans and in Mice.Mol Pharm. 2018 Oct 1;15(10):4589-4598. doi: 10.1021/acs.molpharmaceut.8b00588. Epub 2018 Sep 14. Mol Pharm. 2018. PMID: 30180590
-
Comparative uptake of statins by hepatic organic anion transporting polypeptides.Eur J Pharm Sci. 2025 Jun 1;209:107073. doi: 10.1016/j.ejps.2025.107073. Epub 2025 Mar 17. Eur J Pharm Sci. 2025. PMID: 40107569
-
Erlotinib-A substrate and inhibitor of OATP2B1: pharmacokinetics and CYP3A-mediated metabolism in rSlco2b1-/- and SLCO2B1+/+ rats.Drug Metab Dispos. 2025 May;53(5):100069. doi: 10.1016/j.dmd.2025.100069. Epub 2025 Mar 21. Drug Metab Dispos. 2025. PMID: 40239314
-
Identifying Novel Inhibitors for Hepatic Organic Anion Transporting Polypeptides by Machine Learning-Based Virtual Screening.J Chem Inf Model. 2022 Dec 26;62(24):6323-6335. doi: 10.1021/acs.jcim.1c01460. Epub 2022 Mar 11. J Chem Inf Model. 2022. PMID: 35274943 Free PMC article. Review.
-
Organic anion transporting polypeptide 2B1 - More than a glass-full of drug interactions.Pharmacol Ther. 2019 Apr;196:204-215. doi: 10.1016/j.pharmthera.2018.12.009. Epub 2018 Dec 14. Pharmacol Ther. 2019. PMID: 30557631 Review.
Cited by
-
Impact of rifampicin-inhibitable transport on the liver distribution and tissue kinetics of erlotinib assessed with PET imaging in rats.EJNMMI Res. 2018 Aug 16;8(1):81. doi: 10.1186/s13550-018-0434-0. EJNMMI Res. 2018. PMID: 30116910 Free PMC article.
-
Organic Anion Transporting Polypeptides: Emerging Roles in Cancer Pharmacology.Mol Pharmacol. 2019 May;95(5):490-506. doi: 10.1124/mol.118.114314. Epub 2019 Feb 19. Mol Pharmacol. 2019. PMID: 30782852 Free PMC article. Review.
-
Dissimilar Effect of P-Glycoprotein and Breast Cancer Resistance Protein Inhibition on the Distribution of Erlotinib to the Retina and Brain in Humans and Mice.Mol Pharm. 2023 Nov 6;20(11):5877-5887. doi: 10.1021/acs.molpharmaceut.3c00715. Epub 2023 Oct 26. Mol Pharm. 2023. PMID: 37883694 Free PMC article.
-
Uptake Transporters at the Blood-Brain Barrier and Their Role in Brain Drug Disposition.Pharmaceutics. 2023 Oct 16;15(10):2473. doi: 10.3390/pharmaceutics15102473. Pharmaceutics. 2023. PMID: 37896233 Free PMC article. Review.
-
Impact of Selected Small-Molecule Kinase Inhibitors on Lipid Membranes.Pharmaceuticals (Basel). 2021 Jul 29;14(8):746. doi: 10.3390/ph14080746. Pharmaceuticals (Basel). 2021. PMID: 34451842 Free PMC article.
References
-
- Nies, A.T. , Schwab, M. & Keppler, D. Interplay of conjugating enzymes with OATP uptake transporters and ABCC/MRP efflux pumps in the elimination of drugs. Expert Opin. Drug Metab. Toxicol. 4, 545–568 (2008). - PubMed
-
- Shitara, Y. , Maeda, K. , Ikejiri, K. , Yoshida, K. , Horie, T. & Sugiyama, Y. Clinical significance of organic anion transporting polypeptides (OATPs) in drug disposition: their roles in hepatic clearance and intestinal absorption. Biopharm. Drug Dispos. 34, 45–78 (2013). - PubMed
-
- König, J. , Seithel, A. , Gradhand, U. & Fromm, M.F. Pharmacogenomics of human OATP transporters. Naunyn Schmiedebergs Arch. Pharmacol. 372, 432–443 (2006). - PubMed
-
- König, J. , Müller, F. & Fromm, M.F. Transporters and drug‐drug interactions: important determinants of drug disposition and effects. Pharmacol. Rev. 65, 944–966 (2013). - PubMed
-
- Maeda, K. & Sugiyama, Y. Impact of genetic polymorphisms of transporters on the pharmacokinetic, pharmacodynamic and toxicological properties of anionic drugs. Drug Metab. Pharmacokinet. 23, 223–235 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

