The catalytic mechanism of cyclic GMP-AMP synthase (cGAS) and implications for innate immunity and inhibition
- PMID: 28940468
- PMCID: PMC5699495
- DOI: 10.1002/pro.3304
The catalytic mechanism of cyclic GMP-AMP synthase (cGAS) and implications for innate immunity and inhibition
Abstract
Cyclic GMP-AMP synthase (cGAS) is activated by ds-DNA binding to produce the secondary messenger 2',3'-cGAMP. cGAS is an important control point in the innate immune response; dysregulation of the cGAS pathway is linked to autoimmune diseases while targeted stimulation may be of benefit in immunoncology. We report here the structure of cGAS with dinucleotides and small molecule inhibitors, and kinetic studies of the cGAS mechanism. Our structural work supports the understanding of how ds-DNA activates cGAS, suggesting a site for small molecule binders that may cause cGAS activation at physiological ATP concentrations, and an apparent hotspot for inhibitor binding. Mechanistic studies of cGAS provide the first kinetic constants for 2',3'-cGAMP formation, and interestingly, describe a catalytic mechanism where 2',3'-cGAMP may be a minor product of cGAS compared with linear nucleotides.
Keywords: 2′,3′-cGAMP; OAS1; STING; cGAMP; cGAS; innate immunity.
© 2017 The Authors Protein Science published by Wiley Periodicals, Inc. on behalf of The Protein Society.
Figures

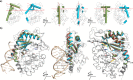
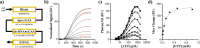

References
-
- Wang Q, Liu X, Zhou Q, Wang C (2015) Cytosolic sensing of aberrant DNA: arming STING on the endoplasmic reticulum. Expert Opin Ther Targets 19:1397–1409. - PubMed
-
- Junt T, Barchet W (2015) Translating nucleic acid‐sensing pathways into therapies. Nat Rev Immunol 15:529–544. - PubMed
-
- Jeremiah N, Neven B, Gentili M, Callebaut I, Maschalidi S, Stolzenberg MC, Goudin N, Fremond ML, Nitschke P, Molina TJ, Blanche S, Picard C, Rice GI, Crow YJ, Manel N, Fischer A, Bader‐Meunier B, Rieux‐Laucat F (2014) Inherited STING‐activating mutation underlies a familial inflammatory syndrome with lupus‐like manifestations. J Clin Invest 124:5516–5520. - PMC - PubMed
-
- Crow YJ (2011) Type I interferonopathies: a novel set of inborn errors of immunity. Ann NY Acad Sci 1238:91–98. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

