The human protein atlas: A spatial map of the human proteome
- PMID: 28940711
- PMCID: PMC5734309
- DOI: 10.1002/pro.3307
The human protein atlas: A spatial map of the human proteome
Abstract
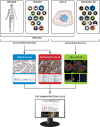
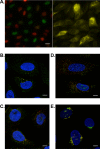
The correct spatial distribution of proteins is vital for their function and often mis-localization or ectopic expression leads to diseases. For more than a decade, the Human Protein Atlas (HPA) has constituted a valuable tool for researchers studying protein localization and expression in human tissues and cells. The centerpiece of the HPA is its unique antibody collection for mapping the entire human proteome by immunohistochemistry and immunocytochemistry. By these approaches, more than 10 million images showing protein expression patterns at a single-cell level were generated and are publicly available at www.proteinatlas.org. The antibody-based approach is combined with transcriptomics data for an overview of global expression profiles. The present article comprehensively describes the HPA database functions and how users can utilize it for their own research as well as discusses the future path of spatial proteomics.
Keywords: antibodies; immunofluorescence; immunohistochemistry; protein expression; single-cell; spatial proteomics; transcriptomics.
© 2017 The Protein Society.
Figures





References
-
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal‐Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LD, Patil AH, Nanjappa V, Radhakrishnan A, Prasad S, Subbannayya T, Raju R, Kumar M, Sreenivasamurthy SK, Marimuthu A, Sathe GJ, Chavan S, Datta KK, Subbannayya Y, Sahu A, Yelamanchi SD, Jayaram S, Rajagopalan P, Sharma J, Murthy KR, Syed N, Goel R, Khan AA, Ahmad S, Dey G, Mudgal K, Chatterjee A, Huang TC, Zhong J, Wu X, Shaw PG, Freed D, Zahari MS, Mukherjee KK, Shankar S, Mahadevan A, Lam H, Mitchell CJ, Shankar SK, Satishchandra P, Schroeder JT, Sirdeshmukh R, Maitra A, Leach SD, Drake CG, Halushka MK, Prasad TS, Hruban RH, Kerr CL, Bader GD, Iacobuzio‐Donahue CA, Gowda H, Pandey A (2014) A draft map of the human proteome. Nature 509:575–581. - PMC - PubMed
-
- Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, Mathieson T, Lemeer S, Schnatbaum K, Reimer U, Wenschuh H, Mollenhauer M, Slotta‐Huspenina J, Boese JH, Bantscheff M, Gerstmair A, Faerber F, Kuster B (2014) Mass‐spectrometry‐based draft of the human proteome. Nature 509:582. - PubMed
-
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F (2015) Tissue‐based map of the human proteome. Science 347:6220. - PubMed
-
- Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM, Bäckström A, Danielsson F, Fagerberg L, Fall J, Gatto L, Gnann C, Hober S, Hjelmare M, Johansson F, Lee S, Lindskog C, Mulder J, Mulvey CM, Nilsson P, Oksvold P, Rockberg J, Schutten R, Schwenk JM, Sivertsson Å, Sjöstedt E, Skogs M, Stadler C, Sullivan DP, Tegel H, Winsnes C, Zhang C, Zwahlen M, Mardinoglu A, Pontén F, von Feilitzen K, Lilley KS, Uhlén M, Lundberg E (2017) A subcellular map of the human proteome. Science 356:eaal3321. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

