Selective pressure for rapid membrane integration constrains the sequence of bacterial outer membrane proteins
- PMID: 28941249
- PMCID: PMC5705044
- DOI: 10.1111/mmi.13845
Selective pressure for rapid membrane integration constrains the sequence of bacterial outer membrane proteins
Abstract
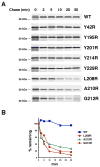
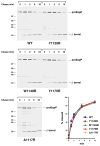
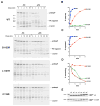
Almost all bacterial outer membrane proteins (OMPs) contain a β barrel domain that serves as a membrane anchor, but the assembly and quality control of these proteins are poorly understood. Here, we show that the introduction of a single lipid-facing arginine residue near the middle of the β barrel of the Escherichia coli OMPs OmpLA and EspP creates an energy barrier that impedes membrane insertion. Although several unintegrated OmpLA mutants remained insertion-competent, they were slowly degraded by the periplasmic protease DegP. Two EspP mutants were also gradually degraded by DegP but were toxic because they first bound to the Bam complex, an essential heteroligomer that catalyzes the membrane insertion of OMPs. Interestingly, another EspP mutant likewise formed a prolonged, deleterious interaction with the Bam complex but was protected from degradation and eventually inserted into the membrane in a native conformation. The different types of interactions between the EspP mutants and the Bam complex that we observed may correspond to distinct stages in OMP assembly. Our results show that sequences that significantly delay assembly are disfavored not only because unintegrated OMPs are subjected to degradation, but also because OMPs that assemble slowly can form dominant-negative interactions with the Bam complex.
© 2017 John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

