Spatial Auxin Signaling Controls Leaf Flattening in Arabidopsis
- PMID: 28943086
- PMCID: PMC6419953
- DOI: 10.1016/j.cub.2017.08.042
Spatial Auxin Signaling Controls Leaf Flattening in Arabidopsis
Abstract
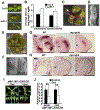
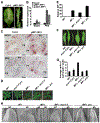
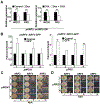
The flattening of leaves to form broad blades is an important adaptation that maximizes photosynthesis. However, the molecular mechanism underlying this process remains unclear. The WUSCHEL-RELATED HOMEOBOX (WOX) genes WOX1 and PRS are expressed in the leaf marginal domain to enable leaf flattening, but the nature of WOX expression establishment remains elusive. Here, we report that adaxial-expressed MONOPTEROS (MP) and abaxial-enriched auxin together act as positional cues for patterning the WOX domain. MP directly binds to the WOX1 and PRS promoters and activates their expression. Furthermore, redundant abaxial-enriched ARF repressors suppress WOX1 and PRS expression, also through direct binding. In particular, we show that ARF2 is redundantly required with ARF3 and ARF4 to maintain the abaxial identity. Taken together, these findings explain how adaxial-abaxial polarity patterns the mediolateral axis and subsequent lateral expansion of leaves.
Keywords: ARF; WOX; auxin; leaf; patterning.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures




Similar articles
-
Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis.Plant Cell. 2012 Feb;24(2):519-35. doi: 10.1105/tpc.111.092858. Epub 2012 Feb 28. Plant Cell. 2012. PMID: 22374393 Free PMC article.
-
A WOX/Auxin Biosynthesis Module Controls Growth to Shape Leaf Form.Curr Biol. 2020 Dec 21;30(24):4857-4868.e6. doi: 10.1016/j.cub.2020.09.037. Epub 2020 Oct 8. Curr Biol. 2020. PMID: 33035489
-
The ASYMMETRIC LEAVES Complex Employs Multiple Modes of Regulation to Affect Adaxial-Abaxial Patterning and Leaf Complexity.Plant Cell. 2015 Dec;27(12):3321-35. doi: 10.1105/tpc.15.00454. Epub 2015 Nov 20. Plant Cell. 2015. PMID: 26589551 Free PMC article.
-
The complex of ASYMMETRIC LEAVES (AS) proteins plays a central role in antagonistic interactions of genes for leaf polarity specification in Arabidopsis.Wiley Interdiscip Rev Dev Biol. 2015 Nov-Dec;4(6):655-71. doi: 10.1002/wdev.196. Epub 2015 Jun 24. Wiley Interdiscip Rev Dev Biol. 2015. PMID: 26108442 Free PMC article. Review.
-
The shady side of leaf development: the role of the REVOLUTA/KANADI1 module in leaf patterning and auxin-mediated growth promotion.Curr Opin Plant Biol. 2017 Feb;35:111-116. doi: 10.1016/j.pbi.2016.11.016. Epub 2016 Dec 3. Curr Opin Plant Biol. 2017. PMID: 27918939 Review.
Cited by
-
Fine mapping of an up-curling leaf locus (BnUC1) in Brassica napus.BMC Plant Biol. 2019 Jul 19;19(1):324. doi: 10.1186/s12870-019-1938-0. BMC Plant Biol. 2019. PMID: 31324149 Free PMC article.
-
The leaf meristem enigma: The relationship between the plate meristem and the marginal meristem.Plant Cell. 2021 Oct 11;33(10):3194-3206. doi: 10.1093/plcell/koab190. Plant Cell. 2021. PMID: 34289073 Free PMC article. Review.
-
Overview of molecular mechanisms of plant leaf development: a systematic review.Front Plant Sci. 2023 Dec 7;14:1293424. doi: 10.3389/fpls.2023.1293424. eCollection 2023. Front Plant Sci. 2023. PMID: 38146273 Free PMC article.
-
Functional analysis of ARF1 from Cymbidium goeringii in IAA response during leaf development.PeerJ. 2022 Mar 10;10:e13077. doi: 10.7717/peerj.13077. eCollection 2022. PeerJ. 2022. PMID: 35291484 Free PMC article.
-
Transgenic tobacco plant overexpressing ginkgo dihydroflavonol 4-reductase gene GbDFR6 exhibits multiple developmental defects.Front Plant Sci. 2022 Dec 14;13:1066736. doi: 10.3389/fpls.2022.1066736. eCollection 2022. Front Plant Sci. 2022. PMID: 36589135 Free PMC article.
References
-
- Waites R, and Hudson A. (1995). phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121, 2143–2154.
-
- Barton MK (2010). Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev Biol 341, 95–113. - PubMed
-
- Bowman JL, and Floyd SK (2008). Patterning and polarity in seed plant shoots. Annu Rev Plant Biol 59, 67–88. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

