CSNK1a1 Regulates PRMT1 to Maintain the Progenitor State in Self-Renewing Somatic Tissue
- PMID: 28943242
- PMCID: PMC5659279
- DOI: 10.1016/j.devcel.2017.08.021
CSNK1a1 Regulates PRMT1 to Maintain the Progenitor State in Self-Renewing Somatic Tissue
Abstract
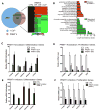
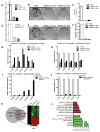
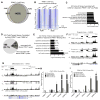
Somatic progenitors sustain tissue self-renewal while suppressing premature differentiation. Protein arginine methyltransferases (PRMTs) affect many processes; however, their role in progenitor function is incompletely understood. PRMT1 was found to be the most highly expressed PRMT in epidermal progenitors and the most downregulated PRMT during differentiation. In targeted mouse knockouts and in long-term regenerated human mosaic epidermis in vivo, epidermal PRMT1 loss abolished progenitor self-renewal and led to premature differentiation. Mass spectrometry of the PRMT1 protein interactome identified the CSNK1a1 kinase, which also proved essential for progenitor maintenance. CSNK1a1 directly bound and phosphorylated PRMT1 to control its genomic targeting to PRMT1-sustained proliferation genes as well as PRMT1-suppressed differentiation genes. Among the latter were GRHL3, whose derepression was required for the premature differentiation seen with PRMT1 and CSNK1a1 loss. Maintenance of the progenitors thus requires cooperation by PRMT1 and CSNK1a1 to sustain proliferation gene expression and suppress premature differentiation driven by GRHL3.
Keywords: CSNK1a1; GRHL3; PRMT1; chromatin; differentiation; epidermis; interactome; keratinocyte; phosphorylation; progenitors.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures




References
-
- An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. - PubMed
-
- Boisvert FM, Côté J, Boulanger MC, Richard S. A proteomic analysis of arginine-methylated protein complexes. Mol Cell Proteomics. 2003;2:1319–1330. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

