An Integrated Outlook on the Metagenome and Metabolome of Intestinal Diseases
- PMID: 28943629
- PMCID: PMC5548254
- DOI: 10.3390/diseases3040341
An Integrated Outlook on the Metagenome and Metabolome of Intestinal Diseases
Abstract
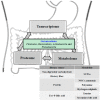
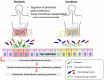
Recently, metagenomics and metabolomics are the two most rapidly advancing "omics" technologies. Metagenomics seeks to characterize the composition of microbial communities, their operations, and their dynamically co-evolving relationships with the habitats they occupy, whereas metabolomics studies unique chemical endpoints (metabolites) that specific cellular processes leave behind. Remarkable progress in DNA sequencing and mass spectrometry technologies has enabled the comprehensive collection of information on the gut microbiome and its metabolome in order to assess the influence of the gut microbiota on host physiology on a whole-systems level. Our gut microbiota, which consists of prokaryotic cells together with its metabolites, creates a unique gut ecosystem together with the host eukaryotic cells. In this review, we will highlight the detailed relationships between gut microbiota and its metabolites on host health and the pathogenesis of various intestinal diseases such as inflammatory bowel disease and colorectal cancer. Therapeutic interventions such as probiotic and prebiotic administrations and fecal microbiota transplantations will also be discussed. We would like to promote this unique biology-wide approach of incorporating metagenome and metabolome information as we believe that this can help us understand the intricate interplay between gut microbiota and host metabolism to a greater extent. This novel integration of microbiome, metatranscriptome, and metabolome information will help us have an improved holistic understanding of the complex mammalian superorganism, thereby allowing us to gain new and unprecedented insights to providing exciting novel therapeutic approaches for optimal intestinal health.
Keywords: colorectal cancer; gut microbiota; inflammatory bowel disease; luminal metabolites; metabolomics; metagenomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Understanding the role of the gut ecosystem in diabetes mellitus.J Diabetes Investig. 2018 Jan;9(1):5-12. doi: 10.1111/jdi.12673. Epub 2017 May 24. J Diabetes Investig. 2018. PMID: 28390093 Free PMC article. Review.
-
Toward the comprehensive understanding of the gut ecosystem via metabolomics-based integrated omics approach.Semin Immunopathol. 2015 Jan;37(1):5-16. doi: 10.1007/s00281-014-0456-2. Epub 2014 Oct 22. Semin Immunopathol. 2015. PMID: 25338280 Review.
-
Metabolome analysis for investigating host-gut microbiota interactions.J Formos Med Assoc. 2019 Mar;118 Suppl 1:S10-S22. doi: 10.1016/j.jfma.2018.09.007. Epub 2018 Sep 27. J Formos Med Assoc. 2019. PMID: 30269936 Review.
-
The human gut microbiome and its dysfunctions through the meta-omics prism.Ann N Y Acad Sci. 2016 May;1372(1):9-19. doi: 10.1111/nyas.13033. Epub 2016 Mar 4. Ann N Y Acad Sci. 2016. PMID: 26945826
-
Gut metabolome meets microbiome: A methodological perspective to understand the relationship between host and microbe.Methods. 2018 Oct 1;149:3-12. doi: 10.1016/j.ymeth.2018.04.029. Epub 2018 Apr 30. Methods. 2018. PMID: 29715508 Review.
Cited by
-
Understanding the role of the gut ecosystem in diabetes mellitus.J Diabetes Investig. 2018 Jan;9(1):5-12. doi: 10.1111/jdi.12673. Epub 2017 May 24. J Diabetes Investig. 2018. PMID: 28390093 Free PMC article. Review.
-
The Application of High-Throughput Technologies for the Study of Microbiome and Cancer.Front Genet. 2021 Jul 28;12:699793. doi: 10.3389/fgene.2021.699793. eCollection 2021. Front Genet. 2021. PMID: 34394190 Free PMC article.
-
Fecal Concentrations of Long-Chain Fatty Acids, Sterols, and Unconjugated Bile Acids in Cats with Chronic Enteropathy.Animals (Basel). 2023 Aug 30;13(17):2753. doi: 10.3390/ani13172753. Animals (Basel). 2023. PMID: 37685017 Free PMC article.
-
Dynamic change of fecal microbiota and metabolomics in a polymicrobial murine sepsis model.Acute Med Surg. 2022 Jun 28;9(1):e770. doi: 10.1002/ams2.770. eCollection 2022 Jan-Dec. Acute Med Surg. 2022. PMID: 35782956 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

