Screening the key genes of hepatocellular adenoma via microarray analysis of DNA expression and methylation profiles
- PMID: 28943905
- PMCID: PMC5605960
- DOI: 10.3892/ol.2017.6673
Screening the key genes of hepatocellular adenoma via microarray analysis of DNA expression and methylation profiles
Abstract
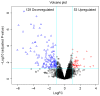
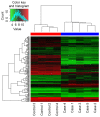
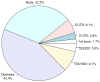
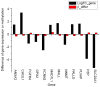
The aim of the present study was to identify the biomarkers involved in the development of hepatocellular adenoma (HCA) through integrated analysis of gene expression and methylation microarray. The microarray dataset GSE7473, containing HNF1α-mutated HCA and their corresponding non-tumor livers, 5 HNF1α-mutated HCA and 4 non-related non-tumor livers, was downloaded from the Gene Expression Omnibus (GEO) database. The DNA methylation profile GSE43091, consisting of 50 HCA and 4 normal liver tissues, was also downloaded from the GEO database. Differentially expressed genes (DEGs) were identified by the limma package of R. A t-test was conducted on the differentially methylated sites. Functional enrichment analysis of DEGs was performed through the Database for Annotation, Visualization and Integrated Analysis. The genes corresponding to the differentially methylated sites were obtained by the annotation files of methylation chip platform. A total of 182 DEGs and 3,902 differentially methylated sites were identified in HCA. In addition, 238 enriched GO terms, including organic acid metabolic process and carboxylic acid metabolic process, and 14 KEGG pathways, including chemical carcinogenesis, were identified. Furthermore, 12 DEGs were identified to contain differentially methylated sites, among which, 8 overlapped genes, including pregnancy zone protein and solute carrier family 22 member 1 (SLC22A1), exhibited inverse associations between gene expression levels and DNA methylation levels. The DNA methylation levels may be potential targets of HCA. The present study revealed that the 8 overlapped genes, including annexin A2, chitinase 3-like 1, fibroblast growth factor receptor 4, mal, T-cell differentiation protein like, palladin, cytoskeletal associated protein, plasmalemma vesicle associated protein and SLC22A1, may be potential therapeutic targets of HCA.
Keywords: DAVID; differentially expressed genes; differentially methylated sites; hepatocellular adenoma.
Figures
Similar articles
-
Identification of potential pathogenic biomarkers in clear cell renal cell carcinoma.Oncol Lett. 2018 Jun;15(6):8491-8499. doi: 10.3892/ol.2018.8398. Epub 2018 Mar 30. Oncol Lett. 2018. PMID: 29805586 Free PMC article.
-
Integrated analysis of genome‑wide gene expression and DNA methylation microarray of diffuse large B‑cell lymphoma with TET mutations.Mol Med Rep. 2017 Oct;16(4):3777-3782. doi: 10.3892/mmr.2017.7058. Epub 2017 Jul 21. Mol Med Rep. 2017. PMID: 28731140 Free PMC article.
-
Identification of potential pathogenic candidates or diagnostic biomarkers in papillary thyroid carcinoma using expression and methylation profiles.Oncol Lett. 2019 Dec;18(6):6670-6678. doi: 10.3892/ol.2019.11059. Epub 2019 Nov 5. Oncol Lett. 2019. PMID: 31814850 Free PMC article.
-
Bioinformatic analysis of the effects and mechanisms of decitabine and cytarabine on acute myeloid leukemia.Mol Med Rep. 2017 Jul;16(1):281-287. doi: 10.3892/mmr.2017.6581. Epub 2017 May 12. Mol Med Rep. 2017. PMID: 28498449 Free PMC article.
-
Comprehensive analysis of DNA methylation gene expression profiles in GEO dataset reveals biomarkers related to malignant transformation of sinonasal inverted papilloma.Discov Oncol. 2024 Mar 1;15(1):53. doi: 10.1007/s12672-024-00903-7. Discov Oncol. 2024. PMID: 38427106 Free PMC article.
Cited by
-
Epigenetic events involved in organic cation transporter 1-dependent impaired response of hepatocellular carcinoma to sorafenib.Br J Pharmacol. 2019 Mar;176(6):787-800. doi: 10.1111/bph.14563. Epub 2019 Feb 11. Br J Pharmacol. 2019. PMID: 30592786 Free PMC article.
-
Current Approaches in the Management of Hepatic Adenomas.J Gastrointest Surg. 2019 Jan;23(1):199-209. doi: 10.1007/s11605-018-3917-4. Epub 2018 Aug 14. J Gastrointest Surg. 2019. PMID: 30109469 Review.
-
Multi-parameter ultrasound based on the logistic regression model in the differential diagnosis of hepatocellular adenoma and focal nodular hyperplasia.World J Gastrointest Oncol. 2019 Dec 15;11(12):1193-1205. doi: 10.4251/wjgo.v11.i12.1193. World J Gastrointest Oncol. 2019. PMID: 31908724 Free PMC article.
-
Expression of microRNA-99a-3p in Prostate Cancer Based on Bioinformatics Data and Meta-Analysis of a Literature Review of 965 Cases.Med Sci Monit. 2018 Jul 12;24:4807-4822. doi: 10.12659/MSM.908057. Med Sci Monit. 2018. PMID: 29997385 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
