Regulation of gene expression is associated with tolerance of the Arctic copepod Calanus glacialis to CO2-acidified sea water
- PMID: 28944006
- PMCID: PMC5606855
- DOI: 10.1002/ece3.3063
Regulation of gene expression is associated with tolerance of the Arctic copepod Calanus glacialis to CO2-acidified sea water
Abstract
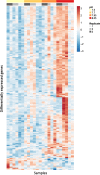
Ocean acidification is the increase in seawater pCO 2 due to the uptake of atmospheric anthropogenic CO 2, with the largest changes predicted to occur in the Arctic seas. For some marine organisms, this change in pCO 2, and associated decrease in pH, represents a climate change-related stressor. In this study, we investigated the gene expression patterns of nauplii of the Arctic copepod Calanus glacialis cultured at low pH levels. We have previously shown that organismal-level performance (development, growth, respiration) of C. glacialis nauplii is unaffected by low pH. Here, we investigated the molecular-level response to lowered pH in order to elucidate the physiological processes involved in this tolerance. Nauplii from wild-caught C. glacialis were cultured at four pH levels (8.05, 7.9, 7.7, 7.5). At stage N6, mRNA was extracted and sequenced using RNA-seq. The physiological functionality of the proteins identified was categorized using Gene Ontology and KEGG pathways. We found that the expression of 151 contigs varied significantly with pH on a continuous scale (93% downregulated with decreasing pH). Gene set enrichment analysis revealed that, of the processes downregulated, many were components of the universal cellular stress response, including DNA repair, redox regulation, protein folding, and proteolysis. Sodium:proton antiporters were among the processes significantly upregulated, indicating that these ion pumps were involved in maintaining cellular pH homeostasis. C. glacialis significantly alters its gene expression at low pH, although they maintain normal larval development. Understanding what confers tolerance to some species will support our ability to predict the effects of future ocean acidification on marine organisms.
Keywords: RNA‐seq; ocean acidification; pH; phenotypic buffering; stress response; transcriptomics.
Figures



References
-
- Aharonovitz, O. , Demaurex, N. , Woodside, M. , & Grinstein, S. (1999). ATP dependence is not an intrinsic property of Na+/H+ exchanger NHE1: requirement for an ancillary factor. American Physiological Society‐ Cell Physiology, 276, 1303–1311. - PubMed
-
- Anderson, K. , Taylor, D. A. , Thompson, E. L. , Melwani, A. R. , Nair, S. V. , & Raftos, D. A. (2015). Meta‐analysis of studies using suppression subtractive hybridization and microarrays to investigate the effects of environmental stress on gene transcription in oysters. PLoS ONE, 10, 1–15. - PMC - PubMed
-
- Arrigo, K. R. , Pabi, S. , Van Dijken, G. L. , & Maslowski, W. (2010). Air‐sea flux of CO2 in the Arctic Ocean, 1998–2003. Journal of Geophysical Research: Biogeosciences, 115, 1998–2003.
LinkOut - more resources
Full Text Sources
Other Literature Sources

