Identification of lineage-specifying cytokines that signal all CD8+-cytotoxic-lineage-fate 'decisions' in the thymus
- PMID: 28945245
- PMCID: PMC5659273
- DOI: 10.1038/ni.3847
Identification of lineage-specifying cytokines that signal all CD8+-cytotoxic-lineage-fate 'decisions' in the thymus
Abstract
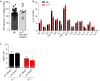
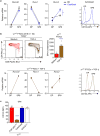
T cell antigen receptor (TCR) signaling in the thymus initiates positive selection, but the CD8+-lineage fate is thought to be induced by cytokines after TCR signaling has ceased, although this remains controversial and unproven. We have identified four cytokines (IL-6, IFN-γ, TSLP and TGF-β) that did not signal via the common γ-chain (γc) receptor but that, like IL-7 and IL-15, induced expression of the lineage-specifying transcription factor Runx3d and signaled the generation of CD8+ T cells. Elimination of in vivo signaling by all six of these 'lineage-specifying cytokines' during positive selection eliminated Runx3d expression and completely abolished the generation of CD8+ single-positive thymocytes. Thus, this study proves that signaling during positive selection by lineage-specifying cytokines is responsible for all CD8+-lineage-fate 'decisions' in the thymus.
Figures





Similar articles
-
Timing and duration of MHC I positive selection signals are adjusted in the thymus to prevent lineage errors.Nat Immunol. 2016 Dec;17(12):1415-1423. doi: 10.1038/ni.3560. Epub 2016 Sep 26. Nat Immunol. 2016. PMID: 27668801 Free PMC article.
-
Conditional deletion of cytokine receptor chains reveals that IL-7 and IL-15 specify CD8 cytotoxic lineage fate in the thymus.J Exp Med. 2012 Nov 19;209(12):2263-76. doi: 10.1084/jem.20121505. Epub 2012 Oct 29. J Exp Med. 2012. PMID: 23109710 Free PMC article.
-
Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice.Nat Rev Immunol. 2008 Oct;8(10):788-801. doi: 10.1038/nri2416. Nat Rev Immunol. 2008. PMID: 18802443 Free PMC article. Review.
-
Overexpression of the Runx3 transcription factor increases the proportion of mature thymocytes of the CD8 single-positive lineage.J Immunol. 2005 Mar 1;174(5):2627-36. doi: 10.4049/jimmunol.174.5.2627. J Immunol. 2005. PMID: 15728469
-
The CD4 Versus CD8 T Cell Fate Decision: A Multiomics-Informed Perspective.Annu Rev Immunol. 2024 Jun;42(1):235-258. doi: 10.1146/annurev-immunol-083122-040929. Epub 2024 Jun 14. Annu Rev Immunol. 2024. PMID: 38271641 Review.
Cited by
-
The Timing and Abundance of IL-2Rβ (CD122) Expression Control Thymic iNKT Cell Generation and NKT1 Subset Differentiation.Front Immunol. 2021 May 14;12:642856. doi: 10.3389/fimmu.2021.642856. eCollection 2021. Front Immunol. 2021. PMID: 34054809 Free PMC article.
-
Direct presentation of inflammation-associated self-antigens by thymic innate-like T cells induces elimination of autoreactive CD8+ thymocytes.Nat Immunol. 2024 Aug;25(8):1367-1382. doi: 10.1038/s41590-024-01899-6. Epub 2024 Jul 11. Nat Immunol. 2024. PMID: 38992254 Free PMC article.
-
Reversal of the T cell immune system reveals the molecular basis for T cell lineage fate determination in the thymus.Nat Immunol. 2022 May;23(5):731-742. doi: 10.1038/s41590-022-01187-1. Epub 2022 Apr 29. Nat Immunol. 2022. PMID: 35523960 Free PMC article.
-
HDAC3 restrains CD8-lineage genes to maintain a bi-potential state in CD4+CD8+ thymocytes for CD4-lineage commitment.Elife. 2019 Jan 18;8:e43821. doi: 10.7554/eLife.43821. Elife. 2019. PMID: 30657451 Free PMC article.
-
CD8 T cell tolerance results from eviction of immature autoreactive cells from the thymus.Science. 2023 Nov 3;382(6670):534-541. doi: 10.1126/science.adh4124. Epub 2023 Nov 2. Science. 2023. PMID: 37917689 Free PMC article.
References
-
- Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. - PubMed
-
- Teh HS, et al. Thymic major histocompatibility complex antigens and the alpha beta T-cell receptor determine the CD4/CD8 phenotype of T cells. Nature. 1988;335:229–233. - PubMed
-
- Brugnera E, et al. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity. 2000;13:59–71. - PubMed
-
- Singer A. New perspectives on a developmental dilemma: the kinetic signaling model and the importance of signal duration for the CD4/CD8 lineage decision. Curr Opin Immunol. 2002;14:207–215. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

