Defective Nucleotide Release by DNA Polymerase β Mutator Variant E288K Is the Basis of Its Low Fidelity
- PMID: 28945359
- PMCID: PMC5654646
- DOI: 10.1021/acs.biochem.7b00869
Defective Nucleotide Release by DNA Polymerase β Mutator Variant E288K Is the Basis of Its Low Fidelity
Abstract
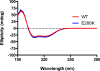
DNA polymerases synthesize new DNA during DNA replication and repair, and their ability to do so faithfully is essential to maintaining genomic integrity. DNA polymerase β (Pol β) functions in base excision repair to fill in single-nucleotide gaps, and variants of Pol β have been associated with cancer. Specifically, the E288K Pol β variant has been found in colon tumors and has been shown to display sequence-specific mutator activity. To probe the mechanism that may underlie E288K's loss of fidelity, a fluorescence resonance energy transfer system that utilizes a fluorophore on the fingers domain of Pol β and a quencher on the DNA substrate was employed. Our results show that E288K utilizes an overall mechanism similar to that of wild type (WT) Pol β when incorporating correct dNTP. However, when inserting the correct dNTP, E288K exhibits a faster rate of closing of the fingers domain combined with a slower rate of nucleotide release compared to those of WT Pol β. We also detect enzyme closure upon mixing with the incorrect dNTP for E288K but not WT Pol β. Taken together, our results suggest that E288K Pol β incorporates all dNTPs more readily than WT because of an inherent defect that results in rapid isomerization of dNTPs within its active site. Structural modeling implies that this inherent defect is due to interaction of E288K with DNA, resulting in a stable closed enzyme structure.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
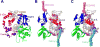







Similar articles
-
The E288K colon tumor variant of DNA polymerase β is a sequence specific mutator.Biochemistry. 2012 Jul 3;51(26):5269-75. doi: 10.1021/bi3003583. Epub 2012 Jun 19. Biochemistry. 2012. PMID: 22650412 Free PMC article.
-
A DNA polymerase beta mutator mutant with reduced nucleotide discrimination and increased protein stability.Biochemistry. 2001 Sep 25;40(38):11372-81. doi: 10.1021/bi010755y. Biochemistry. 2001. PMID: 11560485
-
Hinge residue I174 is critical for proper dNTP selection by DNA polymerase beta.Biochemistry. 2010 Mar 23;49(11):2326-34. doi: 10.1021/bi901735a. Biochemistry. 2010. PMID: 20108981 Free PMC article.
-
Fidelity mechanisms of DNA polymerase beta.Prog Nucleic Acid Res Mol Biol. 2003;73:137-69. doi: 10.1016/s0079-6603(03)01005-5. Prog Nucleic Acid Res Mol Biol. 2003. PMID: 12882517 Review.
-
DNA Polymerase β in the Context of Cancer.Crit Rev Oncog. 2022;27(2):17-33. doi: 10.1615/CritRevOncog.2022043477. Crit Rev Oncog. 2022. PMID: 36734870 Review.
Cited by
-
A pre-catalytic non-covalent step governs DNA polymerase β fidelity.Nucleic Acids Res. 2019 Dec 16;47(22):11839-11849. doi: 10.1093/nar/gkz1076. Nucleic Acids Res. 2019. PMID: 31732732 Free PMC article.
-
I260Q DNA polymerase β highlights precatalytic conformational rearrangements critical for fidelity.Nucleic Acids Res. 2018 Nov 16;46(20):10740-10756. doi: 10.1093/nar/gky825. Nucleic Acids Res. 2018. PMID: 30239932 Free PMC article.
-
Inhibition of DNA Repair in Cancer Therapy: Toward a Multi-Target Approach.Int J Mol Sci. 2020 Sep 12;21(18):6684. doi: 10.3390/ijms21186684. Int J Mol Sci. 2020. PMID: 32932697 Free PMC article. Review.
-
The Impact of SNP-Induced Amino Acid Substitutions L19P and G66R in the dRP-Lyase Domain of Human DNA Polymerase β on Enzyme Activities.Int J Mol Sci. 2024 Apr 10;25(8):4182. doi: 10.3390/ijms25084182. Int J Mol Sci. 2024. PMID: 38673769 Free PMC article.
-
Modifying the Basicity of the dNTP Leaving Group Modulates Precatalytic Conformational Changes of DNA Polymerase β.Biochemistry. 2024 Jun 4;63(11):1412-1422. doi: 10.1021/acs.biochem.4c00065. Epub 2024 May 23. Biochemistry. 2024. PMID: 38780930 Free PMC article.
References
-
- Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

