Thiol-linked alkylation of RNA to assess expression dynamics
- PMID: 28945705
- PMCID: PMC5712218
- DOI: 10.1038/nmeth.4435
Thiol-linked alkylation of RNA to assess expression dynamics
Abstract
Gene expression profiling by high-throughput sequencing reveals qualitative and quantitative changes in RNA species at steady state but obscures the intracellular dynamics of RNA transcription, processing and decay. We developed thiol(SH)-linked alkylation for the metabolic sequencing of RNA (SLAM seq), an orthogonal-chemistry-based RNA sequencing technology that detects 4-thiouridine (s4U) incorporation in RNA species at single-nucleotide resolution. In combination with well-established metabolic RNA labeling protocols and coupled to standard, low-input, high-throughput RNA sequencing methods, SLAM seq enabled rapid access to RNA-polymerase-II-dependent gene expression dynamics in the context of total RNA. We validated the method in mouse embryonic stem cells by showing that the RNA-polymerase-II-dependent transcriptional output scaled with Oct4/Sox2/Nanog-defined enhancer activity, and we provide quantitative and mechanistic evidence for transcript-specific RNA turnover mediated by post-transcriptional gene regulatory pathways initiated by microRNAs and N6-methyladenosine. SLAM seq facilitates the dissection of fundamental mechanisms that control gene expression in an accessible, cost-effective and scalable manner.
Conflict of interest statement
VAH, BR, and SLA declare competing financial interest. A patent application related to this work has been filed.
Figures


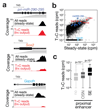

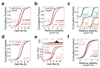
References
-
- Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. - PubMed
-
- Cleary MD, Meiering CD, Jan E, Guymon R, Boothroyd JC. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat Biotechnol. 2005;23:232–237. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

