The promoter region of lapA and its transcriptional regulation by Fis in Pseudomonas putida
- PMID: 28945818
- PMCID: PMC5612765
- DOI: 10.1371/journal.pone.0185482
The promoter region of lapA and its transcriptional regulation by Fis in Pseudomonas putida
Erratum in
-
Correction: The promoter region of lapA and its transcriptional regulation by Fis in Pseudomonas putida.PLoS One. 2018 Jan 30;13(1):e0192336. doi: 10.1371/journal.pone.0192336. eCollection 2018. PLoS One. 2018. PMID: 29381767 Free PMC article.
Abstract
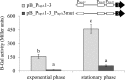
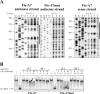
LapA is the biggest protein in Pseudomonas putida and a key factor for biofilm formation. Its importance and posttranslational regulation is rather thoroughly studied but less is known about the transcriptional regulation. Here we give evidence that transcription of lapA in LB-grown bacteria is initiated from six promoters, three of which display moderate RpoS-dependence. The global transcription regulator Fis binds to the lapA promoter area at six positions in vitro, and Fis activates the transcription of lapA while overexpressed in cells. Two of the six Fis binding sites, Fis-A7 and Fis-A5, are necessary for the positive effect of Fis on the transcription of lapA in vivo. Our results indicate that Fis binding to the Fis-A7 site increases the level of transcription from the most distal promoter of lapA, whereas Fis binding to the Fis-A5 site could be important for modifying the promoter area topology.
Conflict of interest statement
Figures









Similar articles
-
Colonization efficiency of Pseudomonas putida is influenced by Fis-controlled transcription of nuoA-N operon.PLoS One. 2018 Aug 2;13(8):e0201841. doi: 10.1371/journal.pone.0201841. eCollection 2018. PLoS One. 2018. PMID: 30071101 Free PMC article.
-
Fis overexpression enhances Pseudomonas putida biofilm formation by regulating the ratio of LapA and LapF.Microbiology (Reading). 2014 Dec;160(Pt 12):2681-2693. doi: 10.1099/mic.0.082503-0. Epub 2014 Sep 24. Microbiology (Reading). 2014. PMID: 25253613
-
Pseudomonas putida Fis binds to the lapF promoter in vitro and represses the expression of LapF.PLoS One. 2014 Dec 29;9(12):e115901. doi: 10.1371/journal.pone.0115901. eCollection 2014. PLoS One. 2014. PMID: 25545773 Free PMC article.
-
Pseudomonas putida Biofilm Depends on the vWFa-Domain of LapA in Peptides-Containing Growth Medium.Int J Mol Sci. 2022 May 24;23(11):5898. doi: 10.3390/ijms23115898. Int J Mol Sci. 2022. PMID: 35682576 Free PMC article.
-
Fis negatively affects binding of Tn4652 transposase by out-competing IHF from the left end of Tn4652.Microbiology (Reading). 2009 Apr;155(Pt 4):1203-1214. doi: 10.1099/mic.0.022830-0. Microbiology (Reading). 2009. PMID: 19332822
Cited by
-
Defining the Genetic Basis of Plant⁻Endophytic Bacteria Interactions.Int J Mol Sci. 2019 Apr 20;20(8):1947. doi: 10.3390/ijms20081947. Int J Mol Sci. 2019. PMID: 31010043 Free PMC article. Review.
-
Catabolism and interactions of syntrophic propionate- and acetate oxidizing microorganisms under mesophilic, high-ammonia conditions.Front Microbiol. 2024 Jun 5;15:1389257. doi: 10.3389/fmicb.2024.1389257. eCollection 2024. Front Microbiol. 2024. PMID: 38933034 Free PMC article.
-
Correction: The promoter region of lapA and its transcriptional regulation by Fis in Pseudomonas putida.PLoS One. 2018 Jan 30;13(1):e0192336. doi: 10.1371/journal.pone.0192336. eCollection 2018. PLoS One. 2018. PMID: 29381767 Free PMC article.
-
Colonization efficiency of Pseudomonas putida is influenced by Fis-controlled transcription of nuoA-N operon.PLoS One. 2018 Aug 2;13(8):e0201841. doi: 10.1371/journal.pone.0201841. eCollection 2018. PLoS One. 2018. PMID: 30071101 Free PMC article.
-
From Input to Output: The Lap/c-di-GMP Biofilm Regulatory Circuit.Annu Rev Microbiol. 2020 Sep 8;74:607-631. doi: 10.1146/annurev-micro-011520-094214. Epub 2020 Jul 20. Annu Rev Microbiol. 2020. PMID: 32689917 Free PMC article. Review.
References
-
- Fazli M, Almblad H, Rybtke ML, Givskov M, Eberl L, Tolker-Nielsen T (2014) Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ Microbiol 16: 1961–1981. doi: 10.1111/1462-2920.12448 - DOI - PubMed
-
- Waite RD, Paccanaro A, Papakonstantinopoulou A, Hurst JM, Saqi M, Littler E, et al. (2006) Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics 7: 162 doi: 10.1186/1471-2164-7-162 - DOI - PMC - PubMed
-
- Moor H, Teppo A, Lahesaare A, Kivisaar M, Teras R (2014) Fis overexpression enhances Pseudomonas putida biofilm formation by regulating the ratio of LapA and LapF. Microbiology 160: 2681–2693. doi: 10.1099/mic.0.082503-0 - DOI - PubMed
-
- Yousef-Coronado F, Travieso ML, Espinosa-Urgel M (2008) Different, overlapping mechanisms for colonization of abiotic and plant surfaces by Pseudomonas putida. FEMS Microbiol Lett 288: 118–124. doi: 10.1111/j.1574-6968.2008.01339.x - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

