Molecular Dynamic Simulation of Space and Earth-Grown Crystal Structures of Thermostable T1 Lipase Geobacillus zalihae Revealed a Better Structure
- PMID: 28946656
- PMCID: PMC6151610
- DOI: 10.3390/molecules22101574
Molecular Dynamic Simulation of Space and Earth-Grown Crystal Structures of Thermostable T1 Lipase Geobacillus zalihae Revealed a Better Structure
Abstract
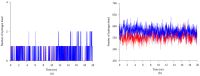
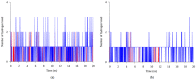
Less sedimentation and convection in a microgravity environment has become a well-suited condition for growing high quality protein crystals. Thermostable T1 lipase derived from bacterium Geobacilluszalihae has been crystallized using the counter diffusion method under space and earth conditions. Preliminary study using YASARA molecular modeling structure program for both structures showed differences in number of hydrogen bond, ionic interaction, and conformation. The space-grown crystal structure contains more hydrogen bonds as compared with the earth-grown crystal structure. A molecular dynamics simulation study was used to provide insight on the fluctuations and conformational changes of both T1 lipase structures. The analysis of root mean square deviation (RMSD), radius of gyration, and root mean square fluctuation (RMSF) showed that space-grown structure is more stable than the earth-grown structure. Space-structure also showed more hydrogen bonds and ion interactions compared to the earth-grown structure. Further analysis also revealed that the space-grown structure has long-lived interactions, hence it is considered as the more stable structure. This study provides the conformational dynamics of T1 lipase crystal structure grown in space and earth condition.
Keywords: Geobacillus zalihae; T1 lipase; hydrogen bond; ion interaction; microgravity; molecular dynamic simulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





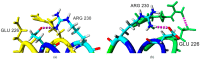
Similar articles
-
Changes of Thermostability, Organic Solvent, and pH Stability in Geobacillus zalihae HT1 and Its Mutant by Calcium Ion.Int J Mol Sci. 2019 May 24;20(10):2561. doi: 10.3390/ijms20102561. Int J Mol Sci. 2019. PMID: 31137725 Free PMC article.
-
A comparative analysis of microgravity and earth grown thermostable T1 lipase crystals using HDPCG apparatus.Protein Pept Lett. 2015;22(2):173-9. doi: 10.2174/0929866521666141019193604. Protein Pept Lett. 2015. PMID: 25329331
-
Ion-Pair Interaction and Hydrogen Bonds as Main Features of Protein Thermostability in Mutated T1 Recombinant Lipase Originating from Geobacillus zalihae.Molecules. 2020 Jul 28;25(15):3430. doi: 10.3390/molecules25153430. Molecules. 2020. PMID: 32731607 Free PMC article.
-
Apocrustacyanin C(1) crystals grown in space and on earth using vapour-diffusion geometry: protein structure refinements and electron-density map comparisons.Acta Crystallogr D Biol Crystallogr. 2003 Jul;59(Pt 7):1117-23. doi: 10.1107/s0907444903007959. Epub 2003 Jun 27. Acta Crystallogr D Biol Crystallogr. 2003. PMID: 12832753 Review.
-
Molecular dynamics of thermoenzymes at high temperature and pressure: a review.Protein J. 2014 Aug;33(4):369-76. doi: 10.1007/s10930-014-9568-8. Protein J. 2014. PMID: 24871480 Review.
Cited by
-
Changes of Thermostability, Organic Solvent, and pH Stability in Geobacillus zalihae HT1 and Its Mutant by Calcium Ion.Int J Mol Sci. 2019 May 24;20(10):2561. doi: 10.3390/ijms20102561. Int J Mol Sci. 2019. PMID: 31137725 Free PMC article.
-
In-silico repurposing of antiviral compounds against Marburg virus: a computational drug discovery approach.In Silico Pharmacol. 2025 Mar 6;13(1):41. doi: 10.1007/s40203-025-00323-7. eCollection 2025. In Silico Pharmacol. 2025. PMID: 40061630
-
CHAPERONg: A tool for automated GROMACS-based molecular dynamics simulations and trajectory analyses.Comput Struct Biotechnol J. 2023 Sep 28;21:4849-4858. doi: 10.1016/j.csbj.2023.09.024. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37854635 Free PMC article.
-
Targeted modulation of MMP9 and GRP78 via molecular interaction and in silico profiling of Curcuma caesia rhizome metabolites: A computational drug discovery approach for cancer therapy.PLoS One. 2025 Jul 18;20(7):e0328509. doi: 10.1371/journal.pone.0328509. eCollection 2025. PLoS One. 2025. PMID: 40679956 Free PMC article.
-
Improved resolution crystal structure of Acanthamoeba actophorin reveals structural plasticity not induced by microgravity.Acta Crystallogr F Struct Biol Commun. 2021 Dec 1;77(Pt 12):452-458. doi: 10.1107/S2053230X21011419. Epub 2021 Nov 11. Acta Crystallogr F Struct Biol Commun. 2021. PMID: 34866600 Free PMC article.
References
-
- Snell E.H., Helliwell J.R. Macromolecular crystallization in microgravity. Rep. Prog. Phys. 2005;68:799–853. doi: 10.1088/0034-4885/68/4/R02. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

