Discrimination of Germline EGFR T790M Mutations in Plasma Cell-Free DNA Allows Study of Prevalence Across 31,414 Cancer Patients
- PMID: 28947568
- PMCID: PMC5712272
- DOI: 10.1158/1078-0432.CCR-17-1745
Discrimination of Germline EGFR T790M Mutations in Plasma Cell-Free DNA Allows Study of Prevalence Across 31,414 Cancer Patients
Abstract
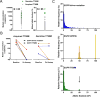
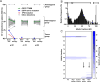
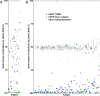
Purpose: Plasma cell-free DNA (cfDNA) analysis is increasingly used clinically for cancer genotyping, but may lead to incidental identification of germline-risk alleles. We studied EGFR T790M mutations in non-small cell lung cancer (NSCLC) toward the aim of discriminating germline and cancer-derived variants within cfDNA.Experimental Design: Patients with EGFR-mutant NSCLC, some with known germline EGFR T790M, underwent plasma genotyping. Separately, deidentified genomic data and buffy coat specimens from a clinical plasma next-generation sequencing (NGS) laboratory were reviewed and tested.Results: In patients with germline T790M mutations, the T790M allelic fraction (AF) in cfDNA approximates 50%, higher than that of EGFR driver mutations. Review of plasma NGS results reveals three groups of variants: a low-AF tumor group, a heterozygous group (∼50% AF), and a homozygous group (∼100% AF). As the EGFR driver mutation AF increases, the distribution of the heterozygous group changes, suggesting increased copy number variation from increased tumor content. Excluding cases with high copy number variation, mutations can be differentiated into somatic variants and incidentally identified germline variants. We then developed a bioinformatic algorithm to distinguish germline and somatic mutations; blinded validation in 21 cases confirmed a 100% positive predictive value for predicting germline T790M. Querying a database of 31,414 patients with plasma NGS, we identified 48 with germline T790M, 43 with nonsquamous NSCLC (P < 0.0001).Conclusions: With appropriate bioinformatics, plasma genotyping can accurately predict the presence of incidentally detected germline risk alleles. This finding in patients indicates a need for genetic counseling and confirmatory germline testing. Clin Cancer Res; 23(23); 7351-9. ©2017 AACR.
©2017 American Association for Cancer Research.
Conflict of interest statement
Odegaard, Fairclough, Nagy, and Lanman are employees of Guardant Health; Nagy and Lanman have ownership interest in Guardant Health. Garber receives research funding or has research collaborations with Myriad Genetics, AstraZeneca, and Ambry and receives consulting fees from Helix. Paweletz received consulting fees from Digital Bioanalytics and honoraria from BioRad. Oxnard received consulting fees from AstraZeneca and Inivata and honoraria from BioRad and Sysmex. All other authors have no potential conflicts of interest to report.
Figures

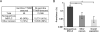
References
-
- Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2(1):82–93. doi: 10.1158/2159-8290.CD-11-0184. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

