Sde2 is an intron-specific pre-mRNA splicing regulator activated by ubiquitin-like processing
- PMID: 28947618
- PMCID: PMC5753039
- DOI: 10.15252/embj.201796751
Sde2 is an intron-specific pre-mRNA splicing regulator activated by ubiquitin-like processing
Abstract
The expression of intron-containing genes in eukaryotes requires generation of protein-coding messenger RNAs (mRNAs) via RNA splicing, whereby the spliceosome removes non-coding introns from pre-mRNAs and joins exons. Spliceosomes must ensure accurate removal of highly diverse introns. We show that Sde2 is a ubiquitin-fold-containing splicing regulator that supports splicing of selected pre-mRNAs in an intron-specific manner in Schizosaccharomyces pombe Both fission yeast and human Sde2 are translated as inactive precursor proteins harbouring the ubiquitin-fold domain linked through an invariant GGKGG motif to a C-terminal domain (referred to as Sde2-C). Precursor processing after the first di-glycine motif by the ubiquitin-specific proteases Ubp5 and Ubp15 generates a short-lived activated Sde2-C fragment with an N-terminal lysine residue, which subsequently gets incorporated into spliceosomes. Absence of Sde2 or defects in Sde2 activation both result in inefficient excision of selected introns from a subset of pre-mRNAs. Sde2 facilitates spliceosomal association of Cactin/Cay1, with a functional link between Sde2 and Cactin further supported by genetic interactions and pre-mRNA splicing assays. These findings suggest that ubiquitin-like processing of Sde2 into a short-lived activated form may function as a checkpoint to ensure proper splicing of certain pre-mRNAs in fission yeast.
Keywords: N‐end rule pathway; deubiquitinating enzymes; intron‐specific pre‐mRNA splicing; telomeric silencing; ubiquitin‐like processing.
© 2017 The Authors.
Figures

Genetic interactors of hub1. hub1‐1 is the temperature‐sensitive hub1(I42S) mutant reported by Yashiroda and Tanaka (2004). A genetic screen was performed with hub1‐1 and the haploid deletion library of non‐essential genes in Schizosaccharomyces pombe. hub1‐1 was synthetically sick with the deletion mutants of given genes including sde2. Among the top hits, genes with relevance to pre‐mRNA splicing are shown.
Confirmation of the negative genetic interaction between hub1‐1 and Δsde2 mutants. sde2 gene, from ATG to the stop codon, was deleted in a S. pombe Δhub1 strain, which was kept viable with a ura4 + marked hub1 expression plasmid with its own promoter and terminator. The resultant strain was transformed with leu2 + marked expression plasmids expressing wild‐type (WT) hub1 or the hub1‐1 mutant. Fivefold serial dilutions of cells from these transformations were spotted on indicated agar plates. Plates were incubated at 25°C. 5‐fluoroorotic acid (FOA) (1 g/l of media) was used to shuffle‐out ura4 + plasmid. Thus, cells growing on −Leu + FOA plates will have hub1 mutants in Δsde2 background.
Alignment of Sde2 protein orthologs from different eukaryotes. Abbreviations used are (respective NCBI protein accession numbers are given in parentheses) as follows: Dh, Debaryomyces hansenii (XP_458854); An, Aspergillus niger (XP_001391007); Sp, Schizosaccharomyces pombe (NP_594019); At, Arabidopsis thaliana (NP_192009); Ce, Caenorhabditis elegans (NP_506378); Ag, Anopheles gambiae (XP_321833); Dm, Drosophila melanogaster (NP_651207); Xl, Xenopus laevis (NP_001084858); Dr, Danio rario (AAH93198); Hs, Homo sapiens (NP_689821). The sequence marked in orange indicates most conserved region in Sde2 orthologs.
Expression of human Sde2 (C1orf55) protein in U2OS cells. Constructs with sequences encoding 3FLAG epitope tag at the N‐terminus of HsSDE2 gene and single MYC epitope tag at its C‐terminus under CMV (cytomegalovirus) promoter were used. With WT Sde2, Sde2‐C would start with lysine (KGG…), whereas with Sde2 GG
A GG mutant, Sde2‐C would start with alanine (AGG…). Asterisk indicates antibody cross reactivity signal. Arrows indicate the HsSde2‐C protein formed after processing of the GGA GG mutant precursor accumulated to higher levels than the protein formed from the wild‐type precursor.

Predicted structure of Schizosaccharomyces pombe Sde2 protein shows the presence of a ubiquitin‐fold (Sde2UBL) and a helical C‐terminal domain (Sde2‐C). The alignment on the left shows the linker GGKGG motif between Sde2UBL and Sde2‐C in Sde2 orthologs. Dh, Debaryomyces hansenii; An, Aspergillus niger; Sp, Schizosaccharomyces pombe; At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Ag, Anopheles gambiae; Dm, Drosophila melanogaster; Xl, Xenopus laevis; Dr, Danio rario; Hs, Homo sapiens.
Expression and processing of Sde2 protein in S. pombe detected by immunoblot analysis (Western blot, WB) using epitope tag‐specific antibodies. Underlined residues mark changes from wild‐type (WT) Sde2. Asterisk indicates antibody cross reactivity signal.
Complementation of S. pombe Δsde2 by GGKGG mutants of Sde2. Constructs are as in (B). Fivefold serial dilution spotting was done on indicated agar plates. Plates were incubated at 30°C and 37°C until growth appeared.
Complementation of S. pombe Δsde2 by Sde2 domains. The experiment is as in (C). Expression constructs encoding Sde2 WT, the processing‐defective mutant sde2(
AA KGG), Sde2UBL, Sde2‐C, and Sde2UBL and Sde2‐C together were used.

Sde2 processing in Schizosaccharomyces pombe. Constructs from Fig 1B were expressed in S. pombe WT, ∆ubp5, ∆ubp15 and ∆ubp5 ∆ubp15 strains. The protein expression was analysed by anti‐FLAG immunoblotting.
Endogenous S. pombe Sde2‐C and the full‐length Sde2 precursor in ∆ubp5 ∆ubp15 strain. Sde2 protein from indicated strains was immunoprecipitated using a polyclonal antibody against recombinant Sde2‐C, followed by Western blot analysis using same antibody.
Sde2 is processed in the nucleus. Anti‐GFP immunoblot analysis of S. pombe expressing Sde2UBL–GFP, and its variants with a nuclear localization signal (NLS) or a nuclear export signal (NES). The non‐nuclear NES version was poorly processed.
Processing of S. pombe Sde2 in bacteria. Expression constructs harbouring indicated cDNAs were co‐transformed in Escherichia coli BL21(DE3) strain. Following protein expression, total cell lysates were processed by immunoblotting using anti‐Sde2‐C antibody. Anti‐HIS immunoblotting was done to monitor expression of the proteases and its mutants.

A USP domain‐containing DUB Ubp16 does not process Sde2. Experiment similar to Fig 2D. Processing of ubiquitin–Sde2‐C fusion by the proteases was used as control. Anti‐HIS immunoblotting was used to monitor expression of the proteases.
Sde2UBL can be replaced with the UBLs ubiquitin and Ned8. Complementation of growth defects of Schizosaccharomyces pombe Δsde2 strain with indicated fusion constructs. All constructs with a C‐terminal 3FLAG tag were expressed under the thiamine‐repressible nmt81 promoter. Expressed proteins are N‐terminal fusions of the ubiquitin‐folds of various UBLs (Ubiquitin/Ubi4, Ned8 or Hub1) with Sde2‐C. The GGKGG motif was kept at the junctions in the fusion proteins. After processing of the UBLs, Sde2‐C would start with a lysine.
Anti‐FLAG immunoblot assay of fusion proteins shown in (B).
Processing of ubiquitin‐ and Ned8–Sde‐C chimeras was not affected in Δubp5 Δubp15 double mutant. Expression of the clones used in (B) and indicated with numerical was monitored by immunoblotting using anti‐FLAG antibody.

Chromosomal 6HA epitope‐tagged sde2 variants. The sde2 promoter is common to all variants.
Western blot with anti‐HA antibody to detect steady‐state levels of wild‐type and mutant Sde2 proteins.
Growth phenotype of strains in (A). Strains with MetSde2‐C proteins, formed either after processing of GG
M GG mutant or translated de novo, show ∆sde2‐like growth defects.Constructs used to monitor steady‐state level and turnover of the proteins. Plasmids contain 3FLAG epitope tags at the C‐termini of Sde2 under nmt81 promoter. Protein turnover assays. The Sde2 variants with C‐terminal 3FLAG tag were expressed in Δsde2 strain from a plasmid under nmt81 promoter. In Sde2 WT, after processing Sde2‐C starts with lysine, in GG
M GG mutant with methionine and Met–Sde2‐C is de novo translated. Total proteins from 1.0 OD600 cells for the given time points were run on SDS–PAGE followed by anti‐FLAG Western blotting.WT Sde2 is stable in the proteasome mutant mts3‐1. Assay is similar to (B). Higher molecular weight adducts detected with anti‐FLAG antibody likely represent ubiquitinated Sde2.
Denaturing Ni‐NTA pull‐down (Pd) of ubiquitin conjugates from Schizosaccharomyces pombe expressing Sde2–3FLAG. Ubiquitination of Sde2 is detected by anti‐FLAG immunoblotting.


Sde2 interacts with the spliceosomal Prp19 complex factors Cdc5, Prp19 and Isy1 in vivo. sde2 gene was 3FLAG‐tagged, and cdc5–6HA, prp19–9MYC and isy1–9MYC genes were tagged chromosomally.
Co‐IP assay to monitor association of the processing‐defective Sde2 with Cdc5. Experiments are performed with proteins expressed at their endogenous levels by using chromosomally tagged strains with given epitopes at the C‐termini of indicated genes. IP was performed using anti‐MYC agarose resins followed by immunoblot assays with anti‐MYC antibody to monitor IP efficiency of Cdc5, and with anti‐HA antibody to monitor Co‐IP of Sde2 versions. Numbers below anti‐HA blot indicate ratio of Sde2 to Cdc5 (HA/MYC) signals obtained from ImageJ quantification of immunoblot signals. Cdc5 association of the processing‐defective Sde2 precursor is diminished to half.
Co‐IP assay to monitor association of the processing‐defective Sde2 with Cwf21. Assay is similar to (B). Cwf21 association of the processing‐defective Sde2 precursor is diminished to one‐third.
Co‐IP assay to monitor association of MetSde2‐C with Cdc5. Assay and quantitation is similar to (B). MetSde2‐C generated from processing of GG
M GG mutant precursor or after translation de novo does not show obvious defects in spliceosomal association.


Analysis of total RNA from Schizosaccharomyces pombe WT and Δsde2 strain using splicing‐sensitive microarray. Microarray heat‐map shows log2 Δsde2/WT ratio of signals for total transcripts (T), intron‐containing transcripts (P) and spliced transcripts (M). 37°C temperature shift was performed for 15 min to monitor early splicing defects. The experiment was repeated with dyes swapped. Yellow colour represents accumulation, black denotes no change, and blue shows reduction of signal in Δsde2.
Semi‐quantitative RT–PCR shows intron‐containing transcripts for psf3, mcs2 and rap1. cDNA prepared from total RNA isolated from S. pombe WT and Δsde2 mutant at 30°C and 37°C was analysed by PCR. The block diagram (not drawn to the scale) shows exons and introns. Arrows mark primers used for amplification. The numbers above introns (shown as lines in block diagrams) mark their lengths. PCR band from genomic DNA (genomic) corresponds to the pre‐mRNA. RT–PCR of act1 (actin) pre‐mRNA is used as control. Asterisks mark intron‐containing transcripts getting enriched in Δsde2.

RT–PCR assays to monitor accumulation of intron‐containing transcripts in Δsde2 and hub1‐1 mutants. Assay is similar to Fig 5B.
Growth phenotypes of chromosomal Sde2 lysine mutants on rich media at 30°C and 37°C.
Association of Cactin/Cay1 with lysine mutants of Sde2‐C. Experiments are performed with proteins expressed from chromosomally tagged strains. IP was performed using anti‐HA beads followed by immunoblot assays with anti‐MYC antibody to monitor Co‐IP of Cay1. Anti‐HA antibody was used to monitor IP efficiency of Sde2 mutants. Numbers indicate ratio of Cay1 to Sde2 (MYC/HA) signals obtained from ImageJ quantification of immunoblots. Results from two independent biological replicate experiments are shown.

Δsde2‐like intron‐specific splicing defect in Δubp5 Δubp15 mutants defective in Sde2 processing. The details are as in Fig 5B.
Intron‐specific splicing defects in processing‐defective and lysine mutants of Sde2. Schizosaccharomyces pombe Δsde2 strain was transformed with vector, Sde2 WT, sde2(
AA KGG) and sde2(GGM GG). The details are as in Fig 5B.

Aberrant overexpression of telomeric transcripts in Sde2 mutant. Expression levels of telomeric transcripts tlh1, tlh2 and SPAC212.06c were quantitated by RT–qPCR assays in WT, and Δsde2 strain at 30°C and 37°C is normalized with act1.
Telomeric silencing assay in Sde2 mutants. ura4 + gene inserted at the telomere in Δsde2 strain (Sugioka‐Sugiyama & Sugiyama, 2011) was used as a marker to assay telomeric silencing on indicated agar plates. Suppression of growth on –Ura plate indicates silencing of the reporter at the telomeres.
Mitotic chromosome stability assay to assess genomic instability in Sde2 mutants. Percentage loss of chromosome 16 in Δsde2 strain (Sugioka‐Sugiyama & Sugiyama, 2011) transformed with Sde2 variants is plotted (n = total number of colonies counted). Processing‐defective and lysine mutants of Sde2 show elevated loss of the chromosome.
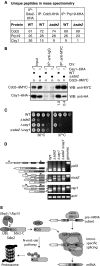
Sde2 Co‐IPs with Cactin in the spliceosome. Cdc5–6HA and Prp19‐6HA complexes were immunoprecipitated using anti‐HA antibody beads from Schizosaccharomyces pombe lysates wild‐type and Δsde2 strains. Co‐IP proteins were analysed by mass spectrometry. The table shows number of unique peptides obtained for each protein in mass spectrometry.
Sde2 facilitates recruitment of Cay1 to the spliceosome. Assay is similar to (A). Lysates from S. pombe wild‐type and Δsde2 strain with Cdc5–9MYC and Cay1–6HA tagged were immunoprecipitated using anti‐MYC antibody beads. Co‐IP of Cay1–6HA was analysed by anti‐HA Western blot. Numbers below anti‐HA blot indicate ratio of Cay1 to Cdc5 (HA/MYC) signals obtained from ImageJ quantification of immunoblot signals.
Sde2 and Cay1 are epistatic. Fivefold serial dilution spotting of indicated mutant strains was done on rich media. Plates were incubated at 30°C and 37°C until growth appeared.
Cay1 and Sde2 knockout strains have similar pre‐mRNA splicing defects. Assay is similar as in Fig 5B.
Schematics for Sde2 function and regulation.
Similar articles
-
Splicing of branchpoint-distant exons is promoted by Cactin, Tls1 and the ubiquitin-fold-activated Sde2.Nucleic Acids Res. 2022 Sep 23;50(17):10000-10014. doi: 10.1093/nar/gkac769. Nucleic Acids Res. 2022. PMID: 36095128 Free PMC article.
-
Intron specificity in pre-mRNA splicing.Curr Genet. 2018 Aug;64(4):777-784. doi: 10.1007/s00294-017-0802-8. Epub 2018 Jan 3. Curr Genet. 2018. PMID: 29299619 Review.
-
The Fission Yeast Pre-mRNA-processing Factor 18 (prp18+) Has Intron-specific Splicing Functions with Links to G1-S Cell Cycle Progression.J Biol Chem. 2016 Dec 30;291(53):27387-27402. doi: 10.1074/jbc.M116.751289. Epub 2016 Nov 15. J Biol Chem. 2016. PMID: 27875300 Free PMC article.
-
Mutant allele of rna14 in fission yeast affects pre-mRNA splicing.J Genet. 2016 Jun;95(2):389-97. doi: 10.1007/s12041-016-0652-z. J Genet. 2016. PMID: 27350684
-
Pre-mRNA splicing in Schizosaccharomyces pombe: regulatory role of a kinase conserved from fission yeast to mammals.Curr Genet. 2003 Feb;42(5):241-51. doi: 10.1007/s00294-002-0355-2. Epub 2002 Dec 13. Curr Genet. 2003. PMID: 12589463 Review.
Cited by
-
Identification of proteins associated with splicing factors Ntr1, Ntr2, Brr2 and Gpl1 in the fission yeast Schizosaccharomyces pombe.Cell Cycle. 2019 Jul;18(14):1532-1536. doi: 10.1080/15384101.2019.1632126. Epub 2019 Jun 20. Cell Cycle. 2019. PMID: 31219728 Free PMC article.
-
A systematic quantitative approach comprehensively defines domain-specific functional pathways linked to Schizosaccharomyces pombe heterochromatin regulation.Nucleic Acids Res. 2024 Dec 11;52(22):13665-13689. doi: 10.1093/nar/gkae1024. Nucleic Acids Res. 2024. PMID: 39565189 Free PMC article.
-
Border cell polarity and collective migration require the spliceosome component Cactin.J Cell Biol. 2022 Jul 4;221(7):e202202146. doi: 10.1083/jcb.202202146. Epub 2022 May 25. J Cell Biol. 2022. PMID: 35612426 Free PMC article.
-
OTULIN Interactome Reveals Immune Response and Autophagy Associated with Tauopathy in a Mouse Model.bioRxiv [Preprint]. 2025 Feb 8:2025.02.07.636114. doi: 10.1101/2025.02.07.636114. bioRxiv. 2025. PMID: 39974971 Free PMC article. Preprint.
-
Splicing of branchpoint-distant exons is promoted by Cactin, Tls1 and the ubiquitin-fold-activated Sde2.Nucleic Acids Res. 2022 Sep 23;50(17):10000-10014. doi: 10.1093/nar/gkac769. Nucleic Acids Res. 2022. PMID: 36095128 Free PMC article.
References
-
- Ast G (2004) How did alternative splicing evolve? Nat Rev Genet 5: 773–782 - PubMed
-
- Bachmair A, Finley D, Varshavsky A (1986) In vivo half‐life of a protein is a function of its amino‐terminal residue. Science 234: 179–186 - PubMed
-
- Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold‐Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C, Landthaler M (2012) The mRNA‐bound proteome and its global occupancy profile on protein‐coding transcripts. Mol Cell 46: 674–690 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

