A dual targeted β-defensin and exome sequencing approach to identify, validate and functionally characterise genes associated with bull fertility
- PMID: 28947819
- PMCID: PMC5613009
- DOI: 10.1038/s41598-017-12498-x
A dual targeted β-defensin and exome sequencing approach to identify, validate and functionally characterise genes associated with bull fertility
Abstract
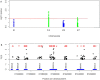
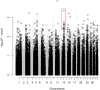
Bovine fertility remains a critical issue underpinning the sustainability of the agricultural sector. Phenotypic records collected on >7,000 bulls used in artificial insemination (AI) were used to identify 160 reliable and divergently fertile bulls for a dual strategy of targeted sequencing (TS) of fertility-related β-defensin genes and whole exome sequencing (WES). A haplotype spanning multiple β-defensin genes and containing 94 SNPs was significantly associated with fertility and functional analysis confirmed that sperm from bulls possessing the haplotype showed significantly enhanced binding to oviductal epithelium. WES of all exons in the genome in 24 bulls of high and low fertility identified 484 additional SNPs significantly associated with fertility. After validation, the most significantly associated SNP was located in the FOXJ3 gene, a transcription factor which regulates sperm function in mice. This study represents the first comprehensive characterisation of genetic variation in bovine β-defensin genes and functional analysis supports a role for β-defensins in regulating bull sperm function. This first application of WES in AI bulls with divergent fertility phenotypes has identified a novel role for the transcription factor FOXJ3 in the regulation of bull fertility. Validated genetic variants associated with bull fertility could prove useful for improving reproductive outcomes in cattle.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
-
- Kearney F. Improving Dairy Herd Fertility through Genetic Selection. Irish Veterinary Journal. 2007;40:376–379.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

