Network reconstruction and systems analysis of plant cell wall deconstruction by Neurospora crassa
- PMID: 28947916
- PMCID: PMC5609067
- DOI: 10.1186/s13068-017-0901-2
Network reconstruction and systems analysis of plant cell wall deconstruction by Neurospora crassa
Abstract
Background: Plant biomass degradation by fungal-derived enzymes is rapidly expanding in economic importance as a clean and efficient source for biofuels. The ability to rationally engineer filamentous fungi would facilitate biotechnological applications for degradation of plant cell wall polysaccharides. However, incomplete knowledge of biomolecular networks responsible for plant cell wall deconstruction impedes experimental efforts in this direction.
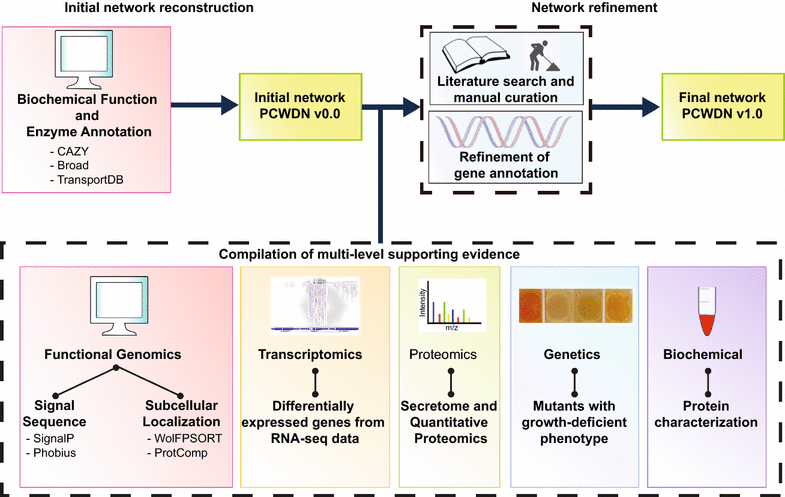
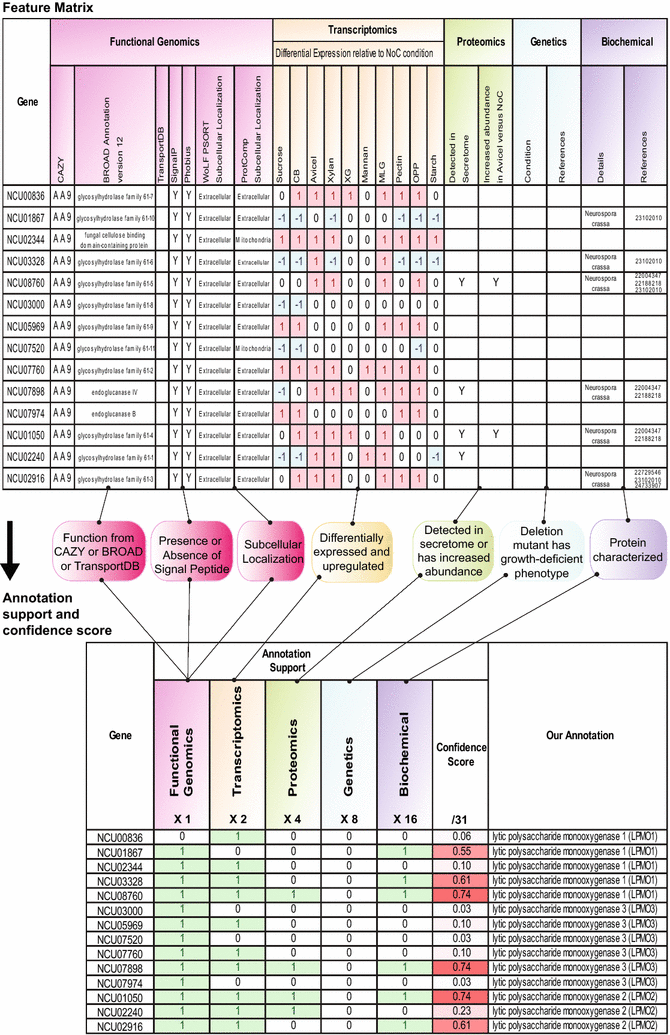
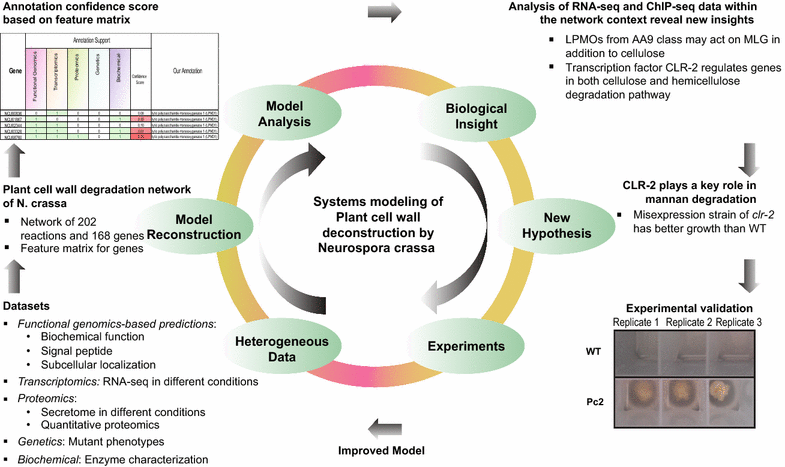
Results: To expand this knowledge base, a detailed network of reactions important for deconstruction of plant cell wall polysaccharides into simple sugars was constructed for the filamentous fungus Neurospora crassa. To reconstruct this network, information was integrated from five heterogeneous data types: functional genomics, transcriptomics, proteomics, genetics, and biochemical characterizations. The combined information was encapsulated into a feature matrix and the evidence weighted to assign annotation confidence scores for each gene within the network. Comparative analyses of RNA-seq and ChIP-seq data shed light on the regulation of the plant cell wall degradation network, leading to a novel hypothesis for degradation of the hemicellulose mannan. The transcription factor CLR-2 was subsequently experimentally shown to play a key role in the mannan degradation pathway of N. crassa.
Conclusions: Here we built a network that serves as a scaffold for integration of diverse experimental datasets. This approach led to the elucidation of regulatory design principles for plant cell wall deconstruction by filamentous fungi and a novel function for the transcription factor CLR-2. This expanding network will aid in efforts to rationally engineer industrially relevant hyper-production strains.
Keywords: Biofuels; CLR-2; Mannan; Network reconstruction; Neurospora crassa; Plant cell wall degradation network; Systems biology; Transcriptional regulatory networks.
Figures






Similar articles
-
The regulatory and transcriptional landscape associated with carbon utilization in a filamentous fungus.Proc Natl Acad Sci U S A. 2020 Mar 17;117(11):6003-6013. doi: 10.1073/pnas.1915611117. Epub 2020 Feb 28. Proc Natl Acad Sci U S A. 2020. PMID: 32111691 Free PMC article.
-
Direct target network of the Neurospora crassa plant cell wall deconstruction regulators CLR-1, CLR-2, and XLR-1.mBio. 2015 Oct 13;6(5):e01452-15. doi: 10.1128/mBio.01452-15. mBio. 2015. PMID: 26463163 Free PMC article.
-
Crosstalk of Cellulose and Mannan Perception Pathways Leads to Inhibition of Cellulase Production in Several Filamentous Fungi.mBio. 2019 Jul 2;10(4):e00277-19. doi: 10.1128/mBio.00277-19. mBio. 2019. PMID: 31266859 Free PMC article.
-
Transcriptional regulation of plant cell wall degradation by filamentous fungi.FEMS Microbiol Rev. 2005 Sep;29(4):719-39. doi: 10.1016/j.femsre.2004.11.006. Epub 2004 Dec 22. FEMS Microbiol Rev. 2005. PMID: 16102600 Review.
-
Neurospora crassa: looking back and looking forward at a model microbe.Am J Bot. 2014 Dec;101(12):2022-35. doi: 10.3732/ajb.1400377. Epub 2014 Nov 16. Am J Bot. 2014. PMID: 25480699 Review.
Cited by
-
Broad Substrate-Specific Phosphorylation Events Are Associated With the Initial Stage of Plant Cell Wall Recognition in Neurospora crassa.Front Microbiol. 2019 Nov 1;10:2317. doi: 10.3389/fmicb.2019.02317. eCollection 2019. Front Microbiol. 2019. PMID: 31736884 Free PMC article.
-
Comparative systems analysis of the secretome of the opportunistic pathogen Aspergillus fumigatus and other Aspergillus species.Sci Rep. 2018 Apr 26;8(1):6617. doi: 10.1038/s41598-018-25016-4. Sci Rep. 2018. PMID: 29700415 Free PMC article.
-
Oxidoreductases and Reactive Oxygen Species in Conversion of Lignocellulosic Biomass.Microbiol Mol Biol Rev. 2018 Sep 26;82(4):e00029-18. doi: 10.1128/MMBR.00029-18. Print 2018 Dec. Microbiol Mol Biol Rev. 2018. PMID: 30257993 Free PMC article. Review.
-
Identification and manipulation of Neurospora crassa genes involved in sensitivity to furfural.Biotechnol Biofuels. 2019 Sep 4;12:210. doi: 10.1186/s13068-019-1550-4. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31508149 Free PMC article.
-
Insights into the Lignocellulose-Degrading Enzyme System Based on the Genome Sequence of Flavodon sp. x-10.Int J Mol Sci. 2025 Jan 21;26(3):866. doi: 10.3390/ijms26030866. Int J Mol Sci. 2025. PMID: 39940637 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

