RETRACTED: Structural analysis of BRCA1 reveals modification hotspot
- PMID: 28948225
- PMCID: PMC5606707
- DOI: 10.1126/sciadv.1701386
RETRACTED: Structural analysis of BRCA1 reveals modification hotspot
Retraction in
-
Editorial Retraction.Sci Adv. 2024 Sep 13;10(37):eads9451. doi: 10.1126/sciadv.ads9451. Epub 2024 Sep 11. Sci Adv. 2024. PMID: 39259782 Free PMC article. No abstract available.
Expression of concern in
-
Editorial expression of concern.Sci Adv. 2024 Mar 15;10(11):eadp1930. doi: 10.1126/sciadv.adp1930. Epub 2024 Mar 13. Sci Adv. 2024. PMID: 38478625 Free PMC article. No abstract available.
Abstract
Cancer cells afflicted with mutations in the breast cancer susceptibility protein (BRCA1) often suffer from increased DNA damage and genomic instability. The precise manner in which physical changes to BRCA1 influence its role in DNA maintenance remains unclear. We used single-particle electron microscopy to study the three-dimensional properties of BRCA1 naturally produced in breast cancer cells. Structural studies revealed new information for full-length BRCA1, engaging its nuclear binding partner, the BRCA1-associated RING domain protein (BARD1). Equally important, we identified a region in mutated BRCA1 that was highly susceptible to ubiquitination. We refer to this site as a modification "hotspot." Ubiquitin adducts in the hotspot region proved to be biochemically reversible. Collectively, we show how key changes to BRCA1 affect its structure-function relationship, and present new insights to potentially modulate mutated BRCA1 in human cancer cells.
Figures



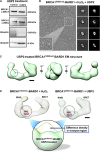
References
-
- Welcsh P. L., Owens K. N., King M.-C., Insights into the functions of BRCA1 and BRCA2. Trends Genet. 16, 69–74 (2000). - PubMed
-
- Rosen E. M., Fan S., Ma Y., BRCA1 regulation of transcription. Cancer Lett. 236, 175–185 (2006). - PubMed
-
- Perou C. M., Molecular stratification of triple-negative breast cancers. Oncologist 15 (suppl. 5), 39–48 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

