Within and beyond the stringent response-RSH and (p)ppGpp in plants
- PMID: 28948393
- PMCID: PMC5633626
- DOI: 10.1007/s00425-017-2780-y
Within and beyond the stringent response-RSH and (p)ppGpp in plants
Abstract
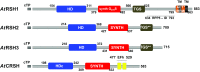
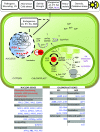
Plant RSH proteins are able to synthetize and/or hydrolyze unusual nucleotides called (p)ppGpp or alarmones. These molecules regulate nuclear and chloroplast transcription, chloroplast translation and plant development and stress response. Homologs of bacterial RelA/SpoT proteins, designated RSH, and products of their activity, (p)ppGpp-guanosine tetra-and pentaphosphates, have been found in algae and higher plants. (p)ppGpp were first identified in bacteria as the effectors of the stringent response, a mechanism that orchestrates pleiotropic adaptations to nutritional deprivation and various stress conditions. (p)ppGpp accumulation in bacteria decreases transcription-with exception to genes that help to withstand or overcome current stressful situations, which are upregulated-and translation as well as DNA replication and eventually reduces metabolism and growth but promotes adaptive responses. In plants, RSH are nuclei-encoded and function in chloroplasts, where alarmones are produced and decrease transcription, translation, hormone, lipid and metabolites accumulation and affect photosynthetic efficiency and eventually plant growth and development. During senescence, alarmones coordinate nutrient remobilization and relocation from vegetative tissues into seeds. Despite the high conservancy of RSH protein domains among bacteria and plants as well as the bacterial origin of plant chloroplasts, in plants, unlike in bacteria, (p)ppGpp promote chloroplast DNA replication and division. Next, (p)ppGpp may also perform their functions in cytoplasm, where they would promote plant growth inhibition. Furthermore, (p)ppGpp accumulation also affects nuclear gene expression, i.a., decreases the level of Arabidopsis defense gene transcripts, and promotes plants susceptibility towards Turnip mosaic virus. In this review, we summarize recent findings that show the importance of RSH and (p)ppGpp in plant growth and development, and open an area of research aiming to understand the function of plant RSH in response to stress.
Keywords: Alarmones; Photosynthesis; Plant growth and development; RelA/SpoT homologs; Senescence; Stress response.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
RSH enzyme diversity for (p)ppGpp metabolism in Phaeodactylum tricornutum and other diatoms.Sci Rep. 2019 Nov 27;9(1):17682. doi: 10.1038/s41598-019-54207-w. Sci Rep. 2019. PMID: 31776430 Free PMC article.
-
Expression profiling of four RelA/SpoT-like proteins, homologues of bacterial stringent factors, in Arabidopsis thaliana.Planta. 2008 Sep;228(4):553-62. doi: 10.1007/s00425-008-0758-5. Epub 2008 Jun 6. Planta. 2008. PMID: 18535838
-
Plastidial (p)ppGpp Synthesis by the Ca2+-Dependent RelA-SpoT Homolog Regulates the Adaptation of Chloroplast Gene Expression to Darkness in Arabidopsis.Plant Cell Physiol. 2021 Feb 4;61(12):2077-2086. doi: 10.1093/pcp/pcaa124. Plant Cell Physiol. 2021. PMID: 33089303
-
Signalling by the global regulatory molecule ppGpp in bacteria and chloroplasts of land plants.Plant Biol (Stuttg). 2011 Sep;13(5):699-709. doi: 10.1111/j.1438-8677.2011.00484.x. Epub 2011 May 31. Plant Biol (Stuttg). 2011. PMID: 21815973 Review.
-
[Plant mechanism of an adaptive stress response homologous to bacterial stringent response].Postepy Biochem. 2006;52(1):94-100. Postepy Biochem. 2006. PMID: 16869307 Review. Polish.
Cited by
-
Identification of a RelA/SpoT Homolog and Its Possible Role in the Accumulation of Astaxanthin in Haematococcus pluvialis.Front Plant Sci. 2022 Feb 9;13:796997. doi: 10.3389/fpls.2022.796997. eCollection 2022. Front Plant Sci. 2022. PMID: 35222463 Free PMC article.
-
Identity and functions of inorganic and inositol polyphosphates in plants.New Phytol. 2020 Jan;225(2):637-652. doi: 10.1111/nph.16129. Epub 2019 Sep 20. New Phytol. 2020. PMID: 31423587 Free PMC article. Review.
-
Retrograde and anterograde signaling in the crosstalk between chloroplast and nucleus.Front Plant Sci. 2022 Sep 2;13:980237. doi: 10.3389/fpls.2022.980237. eCollection 2022. Front Plant Sci. 2022. PMID: 36119624 Free PMC article. Review.
-
RSH enzyme diversity for (p)ppGpp metabolism in Phaeodactylum tricornutum and other diatoms.Sci Rep. 2019 Nov 27;9(1):17682. doi: 10.1038/s41598-019-54207-w. Sci Rep. 2019. PMID: 31776430 Free PMC article.
-
Identification, characterization and expression analysis of wheat RSH family genes under abiotic stress.Front Plant Sci. 2023 Nov 27;14:1283567. doi: 10.3389/fpls.2023.1283567. eCollection 2023. Front Plant Sci. 2023. PMID: 38089805 Free PMC article.
References
-
- Akkaya MS, Breitenberger CA. Light regulation of protein synthesis factor EF-G in pea chloroplasts. Plant Mol Biol. 1992;20:791–800. - PubMed
-
- Anderson PK, Cunningham AA, Patel NG, Morales FJ, Epstein PR, Daszak P. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol Evol. 2004;19:535–544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

