Prevalence and genetic characterisation of respiratory syncytial viruses circulating in Bulgaria during the 2014/15 and 2015/16 winter seasons
- PMID: 28948867
- PMCID: PMC5694888
- DOI: 10.1080/20477724.2017.1375708
Prevalence and genetic characterisation of respiratory syncytial viruses circulating in Bulgaria during the 2014/15 and 2015/16 winter seasons
Abstract
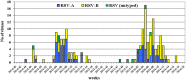
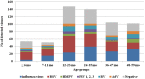
The respiratory syncytial virus (RSV) is a leading cause of acute respiratory illnesses (ARI) in infants and young children. The objectives of this study were to investigate the RSV circulation among children aged <5 years in Bulgaria, to identify the RSV-A and RSV-B genotypes and to perform an amino acid sequence analysis of second hypervariable region (HVR2) of the G gene. During the 2014/15 and 2015/16 winter seasons, nasopharyngeal specimens of 610 children aged <5 years with ARI were tested using Real Time RT-PCR for influenza viruses, RSV, metapneumovirus, parainfluenza viruses, rhinoviruses and adenoviruses. Viral respiratory pathogens were detected in 429 (70%) out of 610 patients examined and RSV was the most frequently identified virus (26%) followed by influenza A(H1N1)pdm09 virus (14%) (p < .05). RSV was the most prevalent pathogen in patients with bronchiolitis (48%) and pneumonia (38%). In the 2014/15 season, RSV-A dominated slightly (53%), while in the next season RSV-B viruses prevailed more strongly (66%). The phylogenetic analysis based on the G gene indicated that all 21 studied RSV-A strains belonged to the ON1 genotype; the vast majority (96%) of the RSV-B strains were classified into BA9 genotype and only one - into BA10 genotype. All Bulgarian RSV-A and RSV-B sequences contained a 72-nt and a 60-nt duplication in the HVR2, respectively. The study showed the leading role of this pathogen as a causative agent of serious respiratory illnesses in early childhood, year-on-year fluctuations in RSV incidence, a shift from RSV-A to RSV-B subgroup dominance and relatively low genetic divergence in the circulating strains.
Keywords: Acute respiratory infection; amino acid substitution; genetic characterization; respiratory syncytial virus.
Figures

References
-
- Collins PL, Karron RA. Respiratory syncytial virus and metapneumovirus In: Knipe DM, Howley PM, editors. Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia: 2013, vol. 1, p. 1086–1123.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
