Stem cell-derived neurons from autistic individuals with SHANK3 mutation show morphogenetic abnormalities during early development
- PMID: 28948968
- PMCID: PMC5822449
- DOI: 10.1038/mp.2017.185
Stem cell-derived neurons from autistic individuals with SHANK3 mutation show morphogenetic abnormalities during early development
Abstract
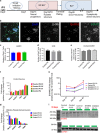
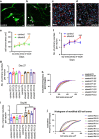
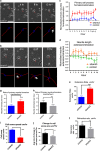
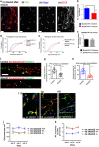
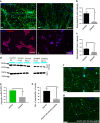
Shank3 is a structural protein found predominantly at the postsynaptic density. Mutations in the SHANK3 gene have been associated with risk for autism spectrum disorder (ASD). We generated induced pluripotent stem cells (iPSCs) from control individuals and from human donors with ASD carrying microdeletions of SHANK3. In addition, we used Zinc finger nucleases to generate isogenic SHANK3 knockout human embryonic stem (ES) cell lines. We differentiated pluripotent cells into either cortical or olfactory placodal neurons. We show that patient-derived placodal neurons make fewer synapses than control cells. Moreover, patient-derived cells display a developmental phenotype: young postmitotic neurons have smaller cell bodies, more extensively branched neurites, and reduced motility compared with controls. These phenotypes were mimicked by SHANK3-edited ES cells and rescued by transduction with a Shank3 expression construct. This developmental phenotype is not observed in the same iPSC lines differentiated into cortical neurons. Therefore, we suggest that SHANK3 has a critical role in neuronal morphogenesis in placodal neurons and that early defects are associated with ASD-associated mutations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
A chimeric mouse model to study human iPSC-derived neurons: the case of a truncating SHANK3 mutation.Sci Rep. 2020 Aug 7;10(1):13315. doi: 10.1038/s41598-020-70056-4. Sci Rep. 2020. PMID: 32769989 Free PMC article.
-
Altered spinogenesis in iPSC-derived cortical neurons from patients with autism carrying de novo SHANK3 mutations.Sci Rep. 2019 Jan 14;9(1):94. doi: 10.1038/s41598-018-36993-x. Sci Rep. 2019. PMID: 30643170 Free PMC article.
-
Shank3 Is Part of a Zinc-Sensitive Signaling System That Regulates Excitatory Synaptic Strength.J Neurosci. 2016 Aug 31;36(35):9124-34. doi: 10.1523/JNEUROSCI.0116-16.2016. J Neurosci. 2016. PMID: 27581454 Free PMC article.
-
Induced pluripotent stem cells as a tool to study brain circuits in autism-related disorders.Stem Cell Res Ther. 2018 Aug 23;9(1):226. doi: 10.1186/s13287-018-0966-2. Stem Cell Res Ther. 2018. PMID: 30139379 Free PMC article. Review.
-
Comparison of SHANK3 deficiency in animal models: phenotypes, treatment strategies, and translational implications.J Neurodev Disord. 2021 Nov 16;13(1):55. doi: 10.1186/s11689-021-09397-8. J Neurodev Disord. 2021. PMID: 34784886 Free PMC article. Review.
Cited by
-
Duplication Versus Deletion Through the Lens of 15q13.3: Clinical and Research Implications of Studying Copy Number Variants Associated with Neuropsychiatric Disorders in Induced Pluripotent Stem Cell-Derived Neurons.Stem Cell Rev Rep. 2023 Apr;19(3):639-650. doi: 10.1007/s12015-022-10475-0. Epub 2022 Nov 12. Stem Cell Rev Rep. 2023. PMID: 36370261 Free PMC article. Review.
-
Insulin-like growth factor 1 partially rescues early developmental defects caused by SHANK2 knockdown in human neurons.Neural Regen Res. 2020 Dec;15(12):2335-2343. doi: 10.4103/1673-5374.285002. Neural Regen Res. 2020. PMID: 32594058 Free PMC article.
-
Human brain organoid model of maternal immune activation identifies radial glia cells as selectively vulnerable.Mol Psychiatry. 2023 Dec;28(12):5077-5089. doi: 10.1038/s41380-023-01997-1. Epub 2023 Mar 6. Mol Psychiatry. 2023. PMID: 36878967 Free PMC article.
-
The Synaptic Gene Study: Design and Methodology to Identify Neurocognitive Markers in Phelan-McDermid Syndrome and NRXN1 Deletions.Front Neurosci. 2022 Feb 18;16:806990. doi: 10.3389/fnins.2022.806990. eCollection 2022. Front Neurosci. 2022. PMID: 35250452 Free PMC article.
-
Distinct effects of interleukin-6 and interferon-γ on differentiating human cortical neurons.Brain Behav Immun. 2022 Jul;103:97-108. doi: 10.1016/j.bbi.2022.04.007. Epub 2022 Apr 13. Brain Behav Immun. 2022. PMID: 35429607 Free PMC article.
References
-
- Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res 2011; 1380: 42–77. - PubMed
-
- LaFlamme B. Genetic modules for autism. Nat Genet 2015; 47: 105–105.
-
- Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol 2009; 19: 231–234. - PubMed
-
- Boeckers TM, Bockmann J, Kreutz MR, Gundelfinger ED. ProSAP/Shank proteins—a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J Neurochem 2002; 81: 903–910. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

