Differential protein-coding gene and long noncoding RNA expression in smoking-related lung squamous cell carcinoma
- PMID: 28949095
- PMCID: PMC5668523
- DOI: 10.1111/1759-7714.12510
Differential protein-coding gene and long noncoding RNA expression in smoking-related lung squamous cell carcinoma
Abstract
Background: Cigarette smoking is one of the greatest preventable risk factors for developing cancer, and most cases of lung squamous cell carcinoma (lung SCC) are associated with smoking. The pathogenesis mechanism of tumor progress is unclear. This study aimed to identify biomarkers in smoking-related lung cancer, including protein-coding gene, long noncoding RNA, and transcription factors.
Methods: We selected and obtained messenger RNA microarray datasets and clinical data from the Gene Expression Omnibus database to identify gene expression altered by cigarette smoking. Integrated bioinformatic analysis was used to clarify biological functions of the identified genes, including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, the construction of a protein-protein interaction network, transcription factor, and statistical analyses. Subsequent quantitative real-time PCR was utilized to verify these bioinformatic analyses.
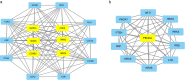
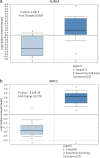
Results: Five hundred and ninety-eight differentially expressed genes and 21 long noncoding RNA were identified in smoking-related lung SCC. GO and KEGG pathway analysis showed that identified genes were enriched in the cancer-related functions and pathways. The protein-protein interaction network revealed seven hub genes identified in lung SCC. Several transcription factors and their binding sites were predicted. The results of real-time quantitative PCR revealed that AURKA and BIRC5 were significantly upregulated and LINC00094 was downregulated in the tumor tissues of smoking patients. Further statistical analysis indicated that dysregulation of AURKA, BIRC5, and LINC00094 indicated poor prognosis in lung SCC.
Conclusion: Protein-coding genes AURKA, BIRC5, and LINC00094 could be biomarkers or therapeutic targets for smoking-related lung SCC.
Keywords: Biomarkers; gene expression profiling; lncRNA; smoking-related lung SCC.
© 2017 The Authors. Thoracic Cancer published by China Lung Oncology Group and John Wiley & Sons Australia, Ltd.
Figures





Similar articles
-
Identification of crucial regulatory relationships between long non-coding RNAs and protein-coding genes in lung squamous cell carcinoma.Mol Cell Probes. 2016 Jun;30(3):146-52. doi: 10.1016/j.mcp.2016.02.009. Epub 2016 Feb 27. Mol Cell Probes. 2016. PMID: 26928440
-
Identification of lung adenocarcinoma biomarkers based on bioinformatic analysis and human samples.Oncol Rep. 2020 May;43(5):1437-1450. doi: 10.3892/or.2020.7526. Epub 2020 Feb 28. Oncol Rep. 2020. PMID: 32323809 Free PMC article.
-
Integrated analysis of long noncoding RNA expression profiles in lymph node metastasis of hepatocellular carcinoma.Gene. 2018 Nov 15;676:47-55. doi: 10.1016/j.gene.2018.07.001. Epub 2018 Jul 4. Gene. 2018. PMID: 29981417
-
Long non-coding RNAs associated with oral squamous cell carcinoma.Eur Rev Med Pharmacol Sci. 2019 Oct;23(20):8888-8896. doi: 10.26355/eurrev_201910_19284. Eur Rev Med Pharmacol Sci. 2019. PMID: 31696475 Review.
-
Global analysis of chromosome 1 genes among patients with lung adenocarcinoma, squamous carcinoma, large-cell carcinoma, small-cell carcinoma, or non-cancer.Cancer Metastasis Rev. 2015 Jun;34(2):249-64. doi: 10.1007/s10555-015-9558-0. Cancer Metastasis Rev. 2015. PMID: 25937073 Review.
Cited by
-
Screening and identification of key biomarkers in lung squamous cell carcinoma by bioinformatics analysis.Oncol Lett. 2019 Nov;18(5):5185-5196. doi: 10.3892/ol.2019.10873. Epub 2019 Sep 16. Oncol Lett. 2019. PMID: 31612029 Free PMC article.
-
Weighted Gene Co-Expression Network Analysis Identifies Critical Genes in the Development of Heart Failure After Acute Myocardial Infarction.Front Genet. 2019 Nov 26;10:1214. doi: 10.3389/fgene.2019.01214. eCollection 2019. Front Genet. 2019. PMID: 31850068 Free PMC article.
-
Lentivirus-mediated shRNA targeting MUTYH inhibits malignant phenotypes of bladder cancer SW780 cells.Onco Targets Ther. 2018 Sep 21;11:6101-6109. doi: 10.2147/OTT.S174223. eCollection 2018. Onco Targets Ther. 2018. PMID: 30275714 Free PMC article.
-
LINC00094/miR-19a-3p/CYP19A1 axis affects the sensitivity of ER positive breast cancer cells to Letrozole through EMT pathway.Aging (Albany NY). 2022 Jun 2;14(11):4755-4768. doi: 10.18632/aging.204110. Epub 2022 Jun 2. Aging (Albany NY). 2022. PMID: 35657638 Free PMC article.
-
ELL associated factor 2 is a potential diagnostic and prognostic indicator: evidence from the in silico and in vitro experiments.Am J Cancer Res. 2023 Jun 15;13(6):2572-2587. eCollection 2023. Am J Cancer Res. 2023. PMID: 37424805 Free PMC article.
References
-
- Chansky K, Detterbeck FC, Nicholson AG et al The IASLC lung cancer staging project: External validation of the revision of the TNM stage groupings in the eighth edition of the TNM classification of lung cancer. J Thorac Oncol 2017; 12: 1109–21. - PubMed
-
- Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol 2016; 893: 1–19. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

