Genomic Landscape of Atypical Adenomatous Hyperplasia Reveals Divergent Modes to Lung Adenocarcinoma
- PMID: 28951454
- PMCID: PMC5774855
- DOI: 10.1158/0008-5472.CAN-17-1605
Genomic Landscape of Atypical Adenomatous Hyperplasia Reveals Divergent Modes to Lung Adenocarcinoma
Abstract
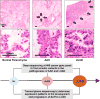
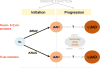
There is a dearth of knowledge about the pathogenesis of premalignant lung lesions, especially for atypical adenomatous hyperplasia (AAH), the only known precursor for the major lung cancer subtype adenocarcinoma (LUAD). In this study, we performed deep DNA and RNA sequencing analyses of a set of AAH, LUAD, and normal tissues. Somatic BRAF variants were found in AAHs from 5 of 22 (23%) patients, 4 of 5 of whom had matched LUAD with driver EGFR mutations. KRAS mutations were present in AAHs from 4 of 22 (18%) of patients. KRAS mutations in AAH were only found in ever-smokers and were exclusive to BRAF-mutant cases. Integrative analysis revealed profiles expressed in KRAS-mutant cases (UBE2C, REL) and BRAF-mutant cases (MAX) of AAH, or common to both sets of cases (suppressed AXL). Gene sets associated with suppressed antitumor (Th1; IL12A, GZMB) and elevated protumor (CCR2, CTLA-4) immune signaling were enriched in AAH development and progression. Our results reveal potentially divergent BRAF or KRAS pathways in AAH as well as immune dysregulation in the pathogenesis of this premalignant lung lesion. Cancer Res; 77(22); 6119-30. ©2017 AACR.
©2017 American Association for Cancer Research.
Conflict of interest statement
Figures
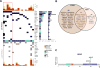



References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. - PubMed
-
- Kadara H, Scheet P, Wistuba II, Spira AE. Early Events in the Molecular Pathogenesis of Lung Cancer. Cancer Prev Res. 2016;9:518–527. - PubMed
-
- Mori M, Rao SK, Popper HH, Cagle PT, Fraire AE. Atypical Adenomatous Hyperplasia of the Lung: A Probable Forerunner in the Development of Adenocarcinoma of the Lung. Mod Pathol. 2001;14:72–84. - PubMed
-
- Sakamoto H, Shimizu J, Horio Y, Ueda R, Takahashi T, Mitsudomi T, et al. Disproportionate representation of KRAS gene mutation in atypical adenomatous hyperplasia, but even distribution of EGFR gene mutation from preinvasive to invasive adenocarcinomas. J Pathol. 2007;212:287–294. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

