Revisiting biomarker discovery by plasma proteomics
- PMID: 28951502
- PMCID: PMC5615924
- DOI: 10.15252/msb.20156297
Revisiting biomarker discovery by plasma proteomics
Abstract
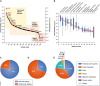
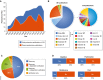
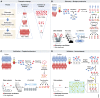
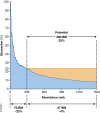
Clinical analysis of blood is the most widespread diagnostic procedure in medicine, and blood biomarkers are used to categorize patients and to support treatment decisions. However, existing biomarkers are far from comprehensive and often lack specificity and new ones are being developed at a very slow rate. As described in this review, mass spectrometry (MS)-based proteomics has become a powerful technology in biological research and it is now poised to allow the characterization of the plasma proteome in great depth. Previous "triangular strategies" aimed at discovering single biomarker candidates in small cohorts, followed by classical immunoassays in much larger validation cohorts. We propose a "rectangular" plasma proteome profiling strategy, in which the proteome patterns of large cohorts are correlated with their phenotypes in health and disease. Translating such concepts into clinical practice will require restructuring several aspects of diagnostic decision-making, and we discuss some first steps in this direction.
Keywords: biomarkers; diagnostic; mass spectrometry; plasma proteomics; systems medicine.
© 2017 The Authors. Published under the terms of the CC BY 4.0 license.
Figures


References
-
- Abbatiello SE, Schilling B, Mani DR, Zimmerman LJ, Hall SC, MacLean B, Albertolle M, Allen S, Burgess M, Cusack MP, Gosh M, Hedrick V, Held JM, Inerowicz HD, Jackson A, Keshishian H, Kinsinger CR, Lyssand J, Makowski L, Mesri M et al (2015) Large‐scale interlaboratory study to develop, analytically validate and apply highly multiplexed, quantitative peptide assays to measure cancer‐relevant proteins in plasma. Mol Cell Proteomics 14: 2357–2374 - PMC - PubMed
-
- Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM et al (2009) Multi‐site assessment of the precision and reproducibility of multiple reaction monitoring‐based measurements of proteins in plasma. Nat Biotechnol 27: 633–641 - PMC - PubMed
-
- Aebersold R, Mann M (2016) Mass‐spectrometric exploration of proteome structure and function. Nature 537: 347–355 - PubMed
-
- Altelaar AF, Heck AJ (2012) Trends in ultrasensitive proteomics. Curr Opin Chem Biol 16: 206–213 - PubMed
-
- Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW (2004) Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti‐Peptide Antibodies (SISCAPA). J Proteome Res 3: 235–244 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

