High-altitude adaptation in humans: from genomics to integrative physiology
- PMID: 28951950
- PMCID: PMC8936998
- DOI: 10.1007/s00109-017-1584-7
High-altitude adaptation in humans: from genomics to integrative physiology
Abstract
About 1.2 to 33% of high-altitude populations suffer from Monge's disease or chronic mountain sickness (CMS). Number of factors such as age, sex, and population of origin (older, male, Andean) contribute to the percentage reported from a variety of samples. It is estimated that there are around 83 million people who live at altitudes > 2500 m worldwide and are at risk for CMS. In this review, we focus on a human "experiment in nature" in various high-altitude locations in the world-namely, Andean, Tibetan, and Ethiopian populations that have lived under chronic hypoxia conditions for thousands of years. We discuss the adaptive as well as mal-adaptive changes at the genomic and physiological levels. Although different genes seem to be involved in adaptation in the three populations, we can observe convergence at genetic and signaling, as well as physiological levels. What is important is that we and others have shown that lessons learned from the genes mined at high altitude can be helpful in better understanding and treating diseases that occur at sea level. We discuss two such examples: EDNRB and SENP1 and their role in cardiac tolerance and in the polycythemic response, respectively.
Keywords: Cardiovascular response; Chronic mountain sickness; Genomics; High-altitude adaptation; Polycythemic response.
Figures
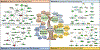



References
-
- Semenza GL (2000) HIF-1 and human disease: one highly involved factor. Genes Dev 14:1983–1991 - PubMed
-
- Semenza GL (2014) Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev Pathol 9:47–71 - PubMed
-
- Nathaniel TI, Williams-Hernandez A, Hunter AL, Liddy C, Peffley DM, Umesiri FE, Imeh-Nathaniel A (2015) Tissue hypoxia during ischemic stroke: adaptive clues from hypoxia-tolerant animal models. Brain Res Bull 114:1–12 - PubMed
-
- Drew KL, Harris MB, LaManna JC, Smith MA, Zhu XW, Ma YL (2004) Hypoxia tolerance in mammalian heterotherms. J Exp Biol 207:3155–3162 - PubMed
-
- Haddad GG (2006) Tolerance to low O2: lessons from inverte-brate genetic models. Exp Physiol 91:277–282 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

