Spaceflight Activates Autophagy Programs and the Proteasome in Mouse Liver
- PMID: 28953266
- PMCID: PMC5666744
- DOI: 10.3390/ijms18102062
Spaceflight Activates Autophagy Programs and the Proteasome in Mouse Liver
Abstract
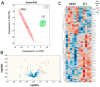
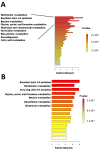
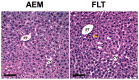
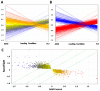
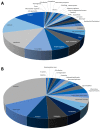
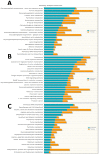
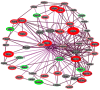
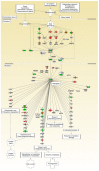
Increased oxidative stress is an unavoidable consequence of exposure to the space environment. Our previous studies showed that mice exposed to space for 13.5 days had decreased glutathione levels, suggesting impairments in oxidative defense. Here we performed unbiased, unsupervised and integrated multi-'omic analyses of metabolomic and transcriptomic datasets from mice flown aboard the Space Shuttle Atlantis. Enrichment analyses of metabolite and gene sets showed significant changes in osmolyte concentrations and pathways related to glycerophospholipid and sphingolipid metabolism, likely consequences of relative dehydration of the spaceflight mice. However, we also found increased enrichment of aminoacyl-tRNA biosynthesis and purine metabolic pathways, concomitant with enrichment of genes associated with autophagy and the ubiquitin-proteasome. When taken together with a downregulation in nuclear factor (erythroid-derived 2)-like 2-mediated signaling, our analyses suggest that decreased hepatic oxidative defense may lead to aberrant tRNA post-translational processing, induction of degradation programs and senescence-associated mitochondrial dysfunction in response to the spaceflight environment.
Keywords: autophagy; metabolomics; proteasome; senescence; spaceflight; tRNA biosynthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Meehan R., Whitson P., Sams C. The role of psychoneuroendocrine factors on spaceflight-induced immunological alterations. J. Leukoc. Biol. 1993;54:236–244. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

