Genome editing reveals a role for OCT4 in human embryogenesis
- PMID: 28953884
- PMCID: PMC5815497
- DOI: 10.1038/nature24033
Genome editing reveals a role for OCT4 in human embryogenesis
Erratum in
-
Erratum: Genome editing reveals a role for OCT4 in human embryogenesis.Nature. 2017 Nov 9;551(7679):256. doi: 10.1038/nature24292. Epub 2017 Oct 4. Nature. 2017. PMID: 28976971
Abstract
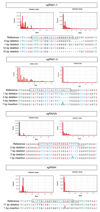
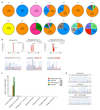
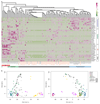
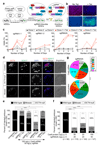
Despite their fundamental biological and clinical importance, the molecular mechanisms that regulate the first cell fate decisions in the human embryo are not well understood. Here we use CRISPR-Cas9-mediated genome editing to investigate the function of the pluripotency transcription factor OCT4 during human embryogenesis. We identified an efficient OCT4-targeting guide RNA using an inducible human embryonic stem cell-based system and microinjection of mouse zygotes. Using these refined methods, we efficiently and specifically targeted the gene encoding OCT4 (POU5F1) in diploid human zygotes and found that blastocyst development was compromised. Transcriptomics analysis revealed that, in POU5F1-null cells, gene expression was downregulated not only for extra-embryonic trophectoderm genes, such as CDX2, but also for regulators of the pluripotent epiblast, including NANOG. By contrast, Pou5f1-null mouse embryos maintained the expression of orthologous genes, and blastocyst development was established, but maintenance was compromised. We conclude that CRISPR-Cas9-mediated genome editing is a powerful method for investigating gene function in the context of human development.
Conflict of interest statement
The authors declare competing financial interests: details are available in the online version of the paper.
Figures








Comment in
-
Take stock of research ethics in human genome editing.Nature. 2017 Sep 20;549(7672):307. doi: 10.1038/549307a. Nature. 2017. PMID: 28933445 No abstract available.
-
At Last: Gene Editing in Human Embryos to Understand Human Development.Cell Stem Cell. 2017 Nov 2;21(5):564-565. doi: 10.1016/j.stem.2017.10.008. Cell Stem Cell. 2017. PMID: 29100008
References
-
- Niakan KK, Eggan K. Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Dev Biol. 2013;375:54–64. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

