Microprocessor Recruitment to Elongating RNA Polymerase II Is Required for Differential Expression of MicroRNAs
- PMID: 28954229
- PMCID: PMC5639929
- DOI: 10.1016/j.celrep.2017.09.010
Microprocessor Recruitment to Elongating RNA Polymerase II Is Required for Differential Expression of MicroRNAs
Abstract
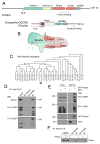
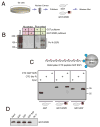
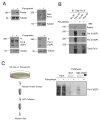
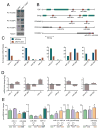
The cellular abundance of mature microRNAs (miRNAs) is dictated by the efficiency of nuclear processing of primary miRNA transcripts (pri-miRNAs) into pre-miRNA intermediates. The Microprocessor complex of Drosha and DGCR8 carries this out, but it has been unclear what controls Microprocessor's differential processing of various pri-miRNAs. Here, we show that Drosophila DGCR8 (Pasha) directly associates with the C-terminal domain of the RNA polymerase II elongation complex when it is phosphorylated by the Cdk9 kinase (pTEFb). When association is blocked by loss of Cdk9 activity, a global change in pri-miRNA processing is detected. Processing of pri-miRNAs with a UGU sequence motif in their apical junction domain increases, while processing of pri-miRNAs lacking this motif decreases. Therefore, phosphorylation of RNA polymerase II recruits Microprocessor for co-transcriptional processing of non-UGU pri-miRNAs that would otherwise be poorly processed. In contrast, UGU-positive pri-miRNAs are robustly processed by Microprocessor independent of RNA polymerase association.
Keywords: DGCR8; Drosophila; RNA polymerase II; microRNA.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Select amino acids in DGCR8 are essential for the UGU-pri-miRNA interaction and processing.Commun Biol. 2020 Jul 3;3(1):344. doi: 10.1038/s42003-020-1071-5. Commun Biol. 2020. PMID: 32620823 Free PMC article.
-
SRSF3 recruits DROSHA to the basal junction of primary microRNAs.RNA. 2018 Jul;24(7):892-898. doi: 10.1261/rna.065862.118. Epub 2018 Apr 3. RNA. 2018. PMID: 29615481 Free PMC article.
-
Microprocessor depends on hemin to recognize the apical loop of primary microRNA.Nucleic Acids Res. 2018 Jun 20;46(11):5726-5736. doi: 10.1093/nar/gky248. Nucleic Acids Res. 2018. PMID: 29750274 Free PMC article.
-
MicroRNA biogenesis: isolation and characterization of the microprocessor complex.Methods Mol Biol. 2006;342:33-47. doi: 10.1385/1-59745-123-1:33. Methods Mol Biol. 2006. PMID: 16957365 Review.
-
Orientation of Human Microprocessor on Primary MicroRNAs.Biochemistry. 2019 Jan 29;58(4):189-198. doi: 10.1021/acs.biochem.8b00944. Epub 2018 Dec 10. Biochemistry. 2019. PMID: 30481000 Review.
Cited by
-
Parameters of clustered suboptimal miRNA biogenesis.Proc Natl Acad Sci U S A. 2023 Oct 10;120(41):e2306727120. doi: 10.1073/pnas.2306727120. Epub 2023 Oct 3. Proc Natl Acad Sci U S A. 2023. PMID: 37788316 Free PMC article.
-
miR-623 suppresses cell proliferation, migration and invasion through direct inhibition of XRCC5 in breast cancer.Aging (Albany NY). 2020 Jun 5;12(11):10246-10258. doi: 10.18632/aging.103182. Epub 2020 Jun 5. Aging (Albany NY). 2020. PMID: 32501811 Free PMC article.
-
Development of novel CDK9 and CYP3A4 inhibitors for cancer therapy through field and computational approaches.Front Chem. 2024 Oct 21;12:1473398. doi: 10.3389/fchem.2024.1473398. eCollection 2024. Front Chem. 2024. PMID: 39498375 Free PMC article.
-
Select amino acids in DGCR8 are essential for the UGU-pri-miRNA interaction and processing.Commun Biol. 2020 Jul 3;3(1):344. doi: 10.1038/s42003-020-1071-5. Commun Biol. 2020. PMID: 32620823 Free PMC article.
-
MicroRNAs: From Mechanism to Organism.Front Cell Dev Biol. 2020 Jun 3;8:409. doi: 10.3389/fcell.2020.00409. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32582699 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

