Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability
- PMID: 28954549
- PMCID: PMC5962345
- DOI: 10.1080/10409238.2017.1380597
Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability
Abstract
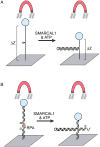
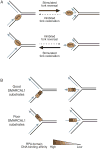
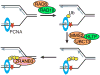
A large number of SNF2 family, DNA and ATP-dependent motor proteins are needed during transcription, DNA replication, and DNA repair to manipulate protein-DNA interactions and change DNA structure. SMARCAL1, ZRANB3, and HLTF are three related members of this family with specialized functions that maintain genome stability during DNA replication. These proteins are recruited to replication forks through protein-protein interactions and bind DNA using both their motor and substrate recognition domains (SRDs). The SRD provides specificity to DNA structures like forks and junctions and confers DNA remodeling activity to the motor domains. Remodeling reactions include fork reversal and branch migration to promote fork stabilization, template switching, and repair. Regulation ensures these powerful activities remain controlled and restricted to damaged replication forks. Inherited mutations in SMARCAL1 cause a severe developmental disorder and mutations in ZRANB3 and HLTF are linked to cancer illustrating the importance of these enzymes in ensuring complete and accurate DNA replication. In this review, we examine how these proteins function, concentrating on their common and unique attributes and regulatory mechanisms.
Keywords: DNA repair; Fork reversal; PCNA; RPA; checkpoint; replication stress.
Figures

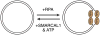



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
