COL4A2 is associated with lacunar ischemic stroke and deep ICH: Meta-analyses among 21,500 cases and 40,600 controls
- PMID: 28954878
- PMCID: PMC5664302
- DOI: 10.1212/WNL.0000000000004560
COL4A2 is associated with lacunar ischemic stroke and deep ICH: Meta-analyses among 21,500 cases and 40,600 controls
Abstract
Objective: To determine whether common variants in familial cerebral small vessel disease (SVD) genes confer risk of sporadic cerebral SVD.
Methods: We meta-analyzed genotype data from individuals of European ancestry to determine associations of common single nucleotide polymorphisms (SNPs) in 6 familial cerebral SVD genes (COL4A1, COL4A2, NOTCH3, HTRA1, TREX1, and CECR1) with intracerebral hemorrhage (ICH) (deep, lobar, all; 1,878 cases, 2,830 controls) and ischemic stroke (IS) (lacunar, cardioembolic, large vessel disease, all; 19,569 cases, 37,853 controls). We applied data quality filters and set statistical significance thresholds accounting for linkage disequilibrium and multiple testing.
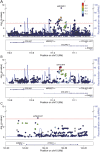
Results: A locus in COL4A2 was associated (significance threshold p < 3.5 × 10-4) with both lacunar IS (lead SNP rs9515201: odds ratio [OR] 1.17, 95% confidence interval [CI] 1.11-1.24, p = 6.62 × 10-8) and deep ICH (lead SNP rs4771674: OR 1.28, 95% CI 1.13-1.44, p = 5.76 × 10-5). A SNP in HTRA1 was associated (significance threshold p < 5.5 × 10-4) with lacunar IS (rs79043147: OR 1.23, 95% CI 1.10-1.37, p = 1.90 × 10-4) and less robustly with deep ICH. There was no clear evidence for association of common variants in either COL4A2 or HTRA1 with non-SVD strokes or in any of the other genes with any stroke phenotype.
Conclusions: These results provide evidence of shared genetic determinants and suggest common pathophysiologic mechanisms of distinct ischemic and hemorrhagic cerebral SVD stroke phenotypes, offering new insights into the causal mechanisms of cerebral SVD.
© 2017 American Academy of Neurology.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- K23 NS064052/NS/NINDS NIH HHS/United States
- K23 NS086873/NS/NINDS NIH HHS/United States
- R01 NS082285/NS/NINDS NIH HHS/United States
- U01 AG009740/AG/NIA NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- R01 NS086905/NS/NINDS NIH HHS/United States
- R01 NS059727/NS/NINDS NIH HHS/United States
- K23 NS100816/NS/NINDS NIH HHS/United States
- KL2 TR001429/TR/NCATS NIH HHS/United States
- R01 HL088521/HL/NHLBI NIH HHS/United States
- U01 NS036695/NS/NINDS NIH HHS/United States
- P30 DK072488/DK/NIDDK NIH HHS/United States
- U01 NS069208/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
