Dysregulation of Alternative Poly-adenylation as a Potential Player in Autism Spectrum Disorder
- PMID: 28955198
- PMCID: PMC5601403
- DOI: 10.3389/fnmol.2017.00279
Dysregulation of Alternative Poly-adenylation as a Potential Player in Autism Spectrum Disorder
Abstract
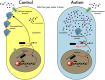
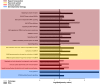
We present here the hypothesis that alternative poly-adenylation (APA) is dysregulated in the brains of individuals affected by Autism Spectrum Disorder (ASD), due to disruptions in the calcium signaling networks. APA, the process of selecting different poly-adenylation sites on the same gene, yielding transcripts with different-length 3' untranslated regions (UTRs), has been documented in different tissues, stages of development and pathologic conditions. Differential use of poly-adenylation sites has been shown to regulate the function, stability, localization and translation efficiency of target RNAs. However, the role of APA remains rather unexplored in neurodevelopmental conditions. In the human brain, where transcripts have the longest 3' UTRs and are thus likely to be under more complex post-transcriptional regulation, erratic APA could be particularly detrimental. In the context of ASD, a condition that affects individuals in markedly different ways and whose symptoms exhibit a spectrum of severity, APA dysregulation could be amplified or dampened depending on the individual and the extent of the effect on specific genes would likely vary with genetic and environmental factors. If this hypothesis is correct, dysregulated APA events might be responsible for certain aspects of the phenotypes associated with ASD. Evidence supporting our hypothesis is derived from standard RNA-seq transcriptomic data but we suggest that future experiments should focus on techniques that probe the actual poly-adenylation site (3' sequencing). To address issues arising from the use of post-mortem tissue and low numbers of heterogeneous samples affected by confounding factors (such as the age, gender and health of the individuals), carefully controlled in vitro systems will be required to model the effect of calcium signaling dysregulation in the ASD brain.
Keywords: RNA–seq; alternative poly-adenylation; autism spectrum disorder; calcium signaling; transcription.
Figures

Similar articles
-
Zika Virus Infection Alters Gene Expression and Poly-Adenylation Patterns in Placental Cells.Pathogens. 2022 Aug 18;11(8):936. doi: 10.3390/pathogens11080936. Pathogens. 2022. PMID: 36015056 Free PMC article.
-
Identification and Characterization of Transcripts Regulated by Circadian Alternative Polyadenylation in Mouse Liver.G3 (Bethesda). 2018 Nov 6;8(11):3539-3548. doi: 10.1534/g3.118.200559. G3 (Bethesda). 2018. PMID: 30181259 Free PMC article.
-
Untranslated Parts of Genes Interpreted: Making Heads or Tails of High-Throughput Transcriptomic Data via Computational Methods: Computational methods to discover and quantify isoforms with alternative untranslated regions.Bioessays. 2017 Dec;39(12). doi: 10.1002/bies.201700090. Epub 2017 Oct 20. Bioessays. 2017. PMID: 29052251 Review.
-
Hypomethylation of miR-142 promoter and upregulation of microRNAs that target the oxytocin receptor gene in the autism prefrontal cortex.Mol Autism. 2015 Aug 14;6:46. doi: 10.1186/s13229-015-0040-1. eCollection 2015. Mol Autism. 2015. PMID: 26273428 Free PMC article.
-
Variation in Gene Expression in Autism Spectrum Disorders: An Extensive Review of Transcriptomic Studies.Front Neurosci. 2017 Jan 5;10:601. doi: 10.3389/fnins.2016.00601. eCollection 2016. Front Neurosci. 2017. PMID: 28105001 Free PMC article. Review.
Cited by
-
RNA Modifications and RNA Metabolism in Neurological Disease Pathogenesis.Int J Mol Sci. 2021 Nov 1;22(21):11870. doi: 10.3390/ijms222111870. Int J Mol Sci. 2021. PMID: 34769301 Free PMC article. Review.
-
Trans-genetic effects of circular RNA expression quantitative trait loci and potential causal mechanisms in autism.Mol Psychiatry. 2022 Nov;27(11):4695-4706. doi: 10.1038/s41380-022-01714-4. Epub 2022 Aug 12. Mol Psychiatry. 2022. PMID: 35962193 Free PMC article.
-
Deciphering the impact of genetic variation on human polyadenylation using APARENT2.Genome Biol. 2022 Nov 5;23(1):232. doi: 10.1186/s13059-022-02799-4. Genome Biol. 2022. PMID: 36335397 Free PMC article.
-
A missense mutation in the CSTF2 gene that impairs the function of the RNA recognition motif and causes defects in 3' end processing is associated with intellectual disability in humans.Nucleic Acids Res. 2020 Sep 25;48(17):9804-9821. doi: 10.1093/nar/gkaa689. Nucleic Acids Res. 2020. PMID: 32816001 Free PMC article.
-
flexiMAP: a regression-based method for discovering differential alternative polyadenylation events in standard RNA-seq data.Bioinformatics. 2021 Jun 16;37(10):1461-1464. doi: 10.1093/bioinformatics/btaa854. Bioinformatics. 2021. PMID: 33051680 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

