The LacI-Family Transcription Factor, RbsR, Is a Pleiotropic Regulator of Motility, Virulence, Siderophore and Antibiotic Production, Gas Vesicle Morphogenesis and Flotation in Serratia
- PMID: 28955306
- PMCID: PMC5601083
- DOI: 10.3389/fmicb.2017.01678
The LacI-Family Transcription Factor, RbsR, Is a Pleiotropic Regulator of Motility, Virulence, Siderophore and Antibiotic Production, Gas Vesicle Morphogenesis and Flotation in Serratia
Abstract
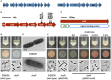
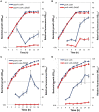
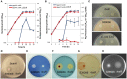
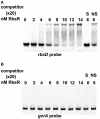
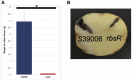
Gas vesicles (GVs) are proteinaceous, gas-filled organelles used by some bacteria to enable upward movement into favorable air/liquid interfaces in aquatic environments. Serratia sp. ATCC39006 (S39006) was the first enterobacterium discovered to produce GVs naturally. The regulation of GV assembly in this host is complex and part of a wider regulatory network affecting various phenotypes, including antibiotic biosynthesis. To identify new regulators of GVs, a comprehensive mutant library containing 71,000 insertion mutants was generated by random transposon mutagenesis and 311 putative GV-defective mutants identified. Three of these mutants were found to have a transposon inserted in a LacI family transcription regulator gene (rbsR) of the putative ribose operon. Each of these rbsR mutants was GV-defective; no GVs were visible by phase contrast microscopy (PCM) or transmission electron microscopy (TEM). GV deficiency was caused by the reduction of gvpA1 and gvrA transcription (the first genes of the two contiguous operons in the GV gene locus). Our results also showed that a mutation in rbsR was highly pleiotropic; the production of two secondary metabolites (carbapenem and prodigiosin antibiotics) was abolished. Interestingly, the intrinsic resistance to the carbapenem antibiotic was not affected by the rbsR mutation. In addition, the production of a siderophore, cellulase and plant virulence was reduced in the mutant, whereas it exhibited increased swimming and swarming motility. The RbsR protein was predicted to bind to regions upstream of at least 18 genes in S39006 including rbsD (the first gene of the ribose operon) and gvrA. Electrophoretic mobility shift assays (EMSA) confirmed that RbsR bound to DNA sequences upstream of rbsD, but not gvrA. The results of this study indicate that RbsR is a global regulator that affects the modulation of GV biogenesis, but also with complex pleiotropic physiological impacts in S39006.
Keywords: Serratia; antibiotics; gas vesicles; gene regulation; motility; ribose operon; virulence.
Figures

Similar articles
-
Environmental potassium regulates bacterial flotation, antibiotic production and turgor pressure in Serratia through the TrkH transporter.Environ Microbiol. 2019 Jul;21(7):2499-2510. doi: 10.1111/1462-2920.14637. Epub 2019 May 13. Environ Microbiol. 2019. PMID: 31012245 Free PMC article.
-
The IclR-family transcriptional regulator XyrR controls flotation, motility, antibiotic production and virulence in Serratia sp. ATCC 39006.Front Microbiol. 2025 Jan 15;15:1500889. doi: 10.3389/fmicb.2024.1500889. eCollection 2024. Front Microbiol. 2025. PMID: 39881986 Free PMC article.
-
The FloR master regulator controls flotation, virulence and antibiotic production in Serratia sp. ATCC 39006.Environ Microbiol. 2020 Jul;22(7):2921-2938. doi: 10.1111/1462-2920.15048. Epub 2020 Jun 14. Environ Microbiol. 2020. PMID: 32352190
-
RNA-seq reveals the RNA binding proteins, Hfq and RsmA, play various roles in virulence, antibiotic production and genomic flux in Serratia sp. ATCC 39006.BMC Genomics. 2013 Nov 22;14(1):822. doi: 10.1186/1471-2164-14-822. BMC Genomics. 2013. PMID: 24267595 Free PMC article.
-
Advances in the application of gas vesicles in medical imaging and disease treatment.J Biol Eng. 2024 Jul 23;18(1):41. doi: 10.1186/s13036-024-00426-3. J Biol Eng. 2024. PMID: 39044273 Free PMC article. Review.
Cited by
-
Genome mining to identify valuable secondary metabolites and their regulation in Actinobacteria from different niches.Arch Microbiol. 2023 Mar 21;205(4):127. doi: 10.1007/s00203-023-03482-3. Arch Microbiol. 2023. PMID: 36944761
-
Improving prodigiosin production by transcription factor engineering and promoter engineering in Serratia marcescens.Front Microbiol. 2022 Aug 3;13:977337. doi: 10.3389/fmicb.2022.977337. eCollection 2022. Front Microbiol. 2022. PMID: 35992721 Free PMC article.
-
RpoS is a pleiotropic regulator of motility, biofilm formation, exoenzymes, siderophore and prodigiosin production, and trade-off during prolonged stationary phase in Serratia marcescens.PLoS One. 2020 Jun 2;15(6):e0232549. doi: 10.1371/journal.pone.0232549. eCollection 2020. PLoS One. 2020. PMID: 32484808 Free PMC article.
-
Improvement of silage characteristics of Lactobacillus salivarius HMC4 and improvement of silage quality of king grass.Front Microbiol. 2024 Dec 11;15:1468577. doi: 10.3389/fmicb.2024.1468577. eCollection 2024. Front Microbiol. 2024. PMID: 39723146 Free PMC article.
-
Environmental potassium regulates bacterial flotation, antibiotic production and turgor pressure in Serratia through the TrkH transporter.Environ Microbiol. 2019 Jul;21(7):2499-2510. doi: 10.1111/1462-2920.14637. Epub 2019 May 13. Environ Microbiol. 2019. PMID: 31012245 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

