A Novel Vector for Construction of Markerless Multicopy Overexpression Transformants in Pichia pastoris
- PMID: 28955309
- PMCID: PMC5601908
- DOI: 10.3389/fmicb.2017.01698
A Novel Vector for Construction of Markerless Multicopy Overexpression Transformants in Pichia pastoris
Abstract
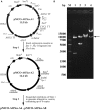
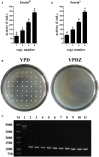
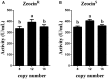
Pichia pastoris is widely used as a platform for heterologous protein expression because of its high volumetric productivity. Multicopy integration of the target gene is commonly used to improve the production of the target protein. Cre/lox recombination system is a powerful tool for the marker rescue during multiple integrations with one selection marker. Here we reported a novel expression vector based on the Cre/lox recombination system for multiple integrations of target gene to construct multicopy expression strain of P. pastoris. PAOX1 promoter was fused to cre to construct a methanol inducible Cre recombinase. The leakage expression of Cre recombinase in Escherichia coli was blocked by introducing the operator gene lacO. The expression vector designed pMCO-AOXα was stable in E. coli and could effectively rescue the Zeocin resistance gene for next round of integration in P. pastoris. Phytase AppA from E. coli was chosen as a reporter gene. Transformants with 2-16 copies of appA were constructed by using a single antibiotic. Expression of appA was gene dosage dependent when <12 copies were integrated. The protein yield increased 4.45-folds when 12 copies of appA were integrated comparing with the single copy integration. Our results showed that pMCO-AOXα was highly effective for rational construction of multicopy transformat in P. pastoris.
Keywords: Pichia pastoris expression system; dosage effect; markerless genetic manipulation; novel expression vector; phytase appA.
Figures




Similar articles
-
[Improving phytase expression by increasing the gene copy number of appA-m in Pichia pastoris].Sheng Wu Gong Cheng Xue Bao. 2006 Jul;22(4):528-33. doi: 10.1016/s1872-2075(06)60041-1. Sheng Wu Gong Cheng Xue Bao. 2006. PMID: 16894882 Chinese.
-
[Overexpression of Escherchia coli phytase with high specific activity].Sheng Wu Gong Cheng Xue Bao. 2004 Jan;20(1):78-84. Sheng Wu Gong Cheng Xue Bao. 2004. PMID: 16108495 Chinese.
-
[Overexpression of Citrobacter braakii phytase with high specific activity in Pichia pastoris].Wei Sheng Wu Xue Bao. 2006 Dec;46(6):945-50. Wei Sheng Wu Xue Bao. 2006. PMID: 17302159 Chinese.
-
Recycling of a selectable marker with a self-excisable plasmid in Pichia pastoris.Sci Rep. 2017 Sep 11;7(1):11113. doi: 10.1038/s41598-017-11494-5. Sci Rep. 2017. PMID: 28894268 Free PMC article.
-
Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production.J Mol Recognit. 2005 Mar-Apr;18(2):119-38. doi: 10.1002/jmr.687. J Mol Recognit. 2005. PMID: 15565717 Review.
Cited by
-
Development of synthetic biology tools to engineer Pichia pastoris as a chassis for the production of natural products.Synth Syst Biotechnol. 2021 May 3;6(2):110-119. doi: 10.1016/j.synbio.2021.04.005. eCollection 2021 Jun. Synth Syst Biotechnol. 2021. PMID: 33997361 Free PMC article. Review.
-
Yeast Expression Systems: Overview and Recent Advances.Mol Biotechnol. 2019 May;61(5):365-384. doi: 10.1007/s12033-019-00164-8. Mol Biotechnol. 2019. PMID: 30805909 Review.
-
Komagataella phaffii as a Platform for Heterologous Expression of Enzymes Used for Industry.Microorganisms. 2024 Feb 7;12(2):346. doi: 10.3390/microorganisms12020346. Microorganisms. 2024. PMID: 38399750 Free PMC article. Review.
-
Current advances of Pichia pastoris as cell factories for production of recombinant proteins.Front Microbiol. 2022 Nov 24;13:1059777. doi: 10.3389/fmicb.2022.1059777. eCollection 2022. Front Microbiol. 2022. PMID: 36504810 Free PMC article. Review.
-
High level production of stable human serum albumin in Pichia pastoris and characterization of the recombinant product.Bioprocess Biosyst Eng. 2022 Feb;45(2):409-424. doi: 10.1007/s00449-021-02670-z. Epub 2022 Jan 9. Bioprocess Biosyst Eng. 2022. PMID: 34999948
References
-
- Çalık P., Ata Ö., Güneş H., Massahi A., Boy E., Keskin A., et al. (2015). Recombinant protein production in Pichia pastoris under glyceraldehyde-3-phosphate dehydrogenase promoter: from carbon source metabolism to bioreactor operation parameters. Biochem. Eng. J. 95, 20–36. 10.1016/j.bej.2014.12.003 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

