Purification and In Vitro Activity of Mitochondria Targeted Nitrogenase Cofactor Maturase NifB
- PMID: 28955359
- PMCID: PMC5601070
- DOI: 10.3389/fpls.2017.01567
Purification and In Vitro Activity of Mitochondria Targeted Nitrogenase Cofactor Maturase NifB
Abstract
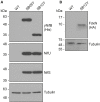
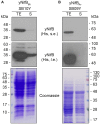
Active NifB is a milestone in the process of engineering nitrogen fixing plants. NifB is an extremely O2-sensitive S-adenosyl methionine (SAM)-radical enzyme that provides the key metal cluster intermediate (NifB-co) for the biosyntheses of the active-site cofactors of all three types of nitrogenases. NifB and NifB-co are unique to diazotrophic organisms. In this work, we have expressed synthetic codon-optimized versions of NifB from the γ-proteobacterium Azotobacter vinelandii and the thermophilic methanogen Methanocaldococcus infernus in Saccharomyces cerevisiae and in Nicotiana benthamiana. NifB proteins were targeted to the mitochondria, where O2 consumption is high and bacterial-like [Fe-S] cluster assembly operates. In yeast, NifB proteins were co-expressed with NifU, NifS, and FdxN proteins that are involved in NifB [Fe-S] cluster assembly and activity. The synthetic version of thermophilic NifB accumulated in soluble form within the yeast cell, while the A. vinelandii version appeared to form aggregates. Similarly, NifB from M. infernus was expressed at higher levels in leaves of Nicotiana benthamiana and accumulated as a soluble protein while A. vinelandii NifB was mainly associated with the non-soluble cell fraction. Soluble M. infernus NifB was purified from aerobically grown yeast and biochemically characterized. The purified protein was functional in the in vitro FeMo-co synthesis assay. This work presents the first active NifB protein purified from a eukaryotic cell, and highlights the importance of screening nif genes from different organisms in order to sort the best candidates to assemble a functional plant nitrogenase.
Keywords: SAM-radical; iron-molybdenum cofactor; mitochondria; nitrogen fixing plants; yeast.
Figures



Similar articles
-
Functional Nitrogenase Cofactor Maturase NifB in Mitochondria and Chloroplasts of Nicotiana benthamiana.mBio. 2022 Jun 28;13(3):e0026822. doi: 10.1128/mbio.00268-22. Epub 2022 Jun 13. mBio. 2022. PMID: 35695456 Free PMC article.
-
Biosynthesis of the nitrogenase active-site cofactor precursor NifB-co in Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 2019 Dec 10;116(50):25078-25086. doi: 10.1073/pnas.1904903116. Epub 2019 Nov 25. Proc Natl Acad Sci U S A. 2019. PMID: 31767756 Free PMC article.
-
Nitrogenase cofactor biosynthesis using proteins produced in mitochondria of Saccharomyces cerevisiae.mBio. 2024 Feb 14;15(2):e0308823. doi: 10.1128/mbio.03088-23. Epub 2023 Dec 21. mBio. 2024. PMID: 38126768 Free PMC article.
-
Structural Insights and Mechanistic Understanding of Iron-Molybdenum Cofactor Biosynthesis by NifB in Nitrogenase Assembly Process.Mol Cells. 2023 Dec 31;46(12):736-742. doi: 10.14348/molcells.2023.0140. Epub 2023 Nov 29. Mol Cells. 2023. PMID: 38052488 Free PMC article. Review.
-
Radical SAM-dependent formation of a nitrogenase cofactor core on NifB.J Inorg Biochem. 2022 Aug;233:111837. doi: 10.1016/j.jinorgbio.2022.111837. Epub 2022 Apr 20. J Inorg Biochem. 2022. PMID: 35550498 Free PMC article. Review.
Cited by
-
Plant expression of NifD protein variants resistant to mitochondrial degradation.Proc Natl Acad Sci U S A. 2020 Sep 15;117(37):23165-23173. doi: 10.1073/pnas.2002365117. Epub 2020 Aug 31. Proc Natl Acad Sci U S A. 2020. PMID: 32868448 Free PMC article.
-
Biosynthesis of cofactor-activatable iron-only nitrogenase in Saccharomyces cerevisiae.Microb Biotechnol. 2021 May;14(3):1073-1083. doi: 10.1111/1751-7915.13758. Epub 2021 Jan 28. Microb Biotechnol. 2021. PMID: 33507628 Free PMC article.
-
Diversity and Functional Analysis of the FeMo-Cofactor Maturase NifB.Front Plant Sci. 2017 Nov 14;8:1947. doi: 10.3389/fpls.2017.01947. eCollection 2017. Front Plant Sci. 2017. PMID: 29250084 Free PMC article.
-
Are we there yet? The long walk towards the development of efficient symbiotic associations between nitrogen-fixing bacteria and non-leguminous crops.BMC Biol. 2019 Dec 3;17(1):99. doi: 10.1186/s12915-019-0710-0. BMC Biol. 2019. PMID: 31796086 Free PMC article. Review.
-
A Synthetic Biology Workflow Reveals Variation in Processing and Solubility of Nitrogenase Proteins Targeted to Plant Mitochondria, and Differing Tolerance of Targeting Sequences in a Bacterial Nitrogenase Assay.Front Plant Sci. 2020 Sep 10;11:552160. doi: 10.3389/fpls.2020.552160. eCollection 2020. Front Plant Sci. 2020. PMID: 33013970 Free PMC article.
References
-
- Bishop P. E., Joerger R. D. (1990). Genetics and molecular biology of alternative nitrogen fixation systems. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41 109–125. 10.1146/annurev.pp.41.060190.000545 - DOI
-
- Borlaug N. E. (2002). Feeding a world of 10 billion people: the miracle ahead. Vitr. Cell. Dev. Biol. Plant 38 221–228. 10.1079/IVP2001279 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

