ABSCISIC ACID INSENSITIVE3 Is Involved in Cold Response and Freezing Tolerance Regulation in Physcomitrella patens
- PMID: 28955377
- PMCID: PMC5601040
- DOI: 10.3389/fpls.2017.01599
ABSCISIC ACID INSENSITIVE3 Is Involved in Cold Response and Freezing Tolerance Regulation in Physcomitrella patens
Abstract
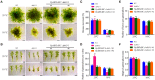
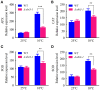
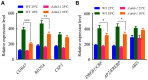
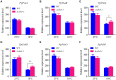
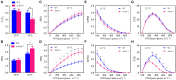
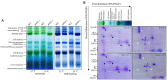
Synopsis This work demonstrates that PpABI3 contributes to freezing tolerance regulation in Physcomitrella patens. Transcription factor ABSCISIC ACID INSENSITIVE3 (ABI3) is known to play a major role in regulating seed dormancy, germination, seedling development as well as stress responses. ABI3 is conserved among land plants; however, its roles in non-seed plants under stress conditions have not been well characterized. In this study, we report that ABI3 is involved in freezing tolerance regulation during cold acclimation at least in part through ABA signaling pathway in moss Physcomitrella patens (P. patens). Deletion of PpABI3 (Δabi3-1) compromises the induction of genes related to cold response and antioxidative protection, resulting in reduced accumulation of cryoprotectants and antioxidants. In addition, photosystem II (PSII) activity is repressed in Δabi3-1 during cold acclimation partially due to alternations of photosynthetic protein complexes compositions. The gametophyte of Δabi3-1 displays severe growth inhibition and developmental deficiency under low temperature condition, while two independent complementary lines display phenotypes similar to that of wild-type P. patens (WT). Furthermore, the freezing tolerance of Δabi3-1 was significantly affected by deletion of PpABI3. These data revealed that PpABI3 plays an important role in low temperature response and freezing tolerance in P. patens.
Keywords: ABA; ABI3; Physcomitrella patens; cold acclimation; freezing tolerance; photosynthetic protein complexes.
Figures



Similar articles
-
Regulatory Mechanism of ABA and ABI3 on Vegetative Development in the Moss Physcomitrella patens.Int J Mol Sci. 2018 Sep 12;19(9):2728. doi: 10.3390/ijms19092728. Int J Mol Sci. 2018. PMID: 30213069 Free PMC article.
-
Large-scale proteome analysis of abscisic acid and ABSCISIC ACID INSENSITIVE3-dependent proteins related to desiccation tolerance in Physcomitrella patens.Biochem Biophys Res Commun. 2016 Mar 18;471(4):589-95. doi: 10.1016/j.bbrc.2016.02.024. Epub 2016 Feb 8. Biochem Biophys Res Commun. 2016. PMID: 26869511
-
Cold acclimation in the moss Physcomitrella patens involves abscisic acid-dependent signaling.J Plant Physiol. 2012 Jan 15;169(2):137-45. doi: 10.1016/j.jplph.2011.08.004. Epub 2011 Sep 28. J Plant Physiol. 2012. PMID: 21958596
-
Abscisic acid-induced freezing tolerance in the moss Physcomitrella patens is accompanied by increased expression of stress-related genes.J Plant Physiol. 2003 May;160(5):475-83. doi: 10.1078/0176-1617-00888. J Plant Physiol. 2003. PMID: 12806775
-
Physcomitrella patens: mosses enter the genomic age.Curr Opin Plant Biol. 2007 Apr;10(2):182-9. doi: 10.1016/j.pbi.2007.01.005. Epub 2007 Feb 8. Curr Opin Plant Biol. 2007. PMID: 17291824 Review.
Cited by
-
Abscisic acid enhances non-photochemical quenching through SnRK2 and ABI3 in Physcomitrium patens.J Plant Res. 2025 Jul;138(4):625-636. doi: 10.1007/s10265-025-01627-7. Epub 2025 Apr 7. J Plant Res. 2025. PMID: 40192897
-
Chilling stress drives organ-specific transcriptional cascades and dampens diurnal oscillation in tomato.Hortic Res. 2023 Jul 11;10(8):uhad137. doi: 10.1093/hr/uhad137. eCollection 2023 Aug. Hortic Res. 2023. PMID: 37564269 Free PMC article.
-
Inositol Improves Cold Tolerance Through Inhibiting CBL1 and Increasing Ca2+ Influx in Rapeseed (Brassica napus L.).Front Plant Sci. 2022 Mar 17;13:775692. doi: 10.3389/fpls.2022.775692. eCollection 2022. Front Plant Sci. 2022. PMID: 35371155 Free PMC article.
-
Pseudocrossidium replicatum (Taylor) R.H. Zander is a fully desiccation-tolerant moss that expresses an inducible molecular mechanism in response to severe abiotic stress.Plant Mol Biol. 2021 Nov;107(4-5):387-404. doi: 10.1007/s11103-021-01167-3. Epub 2021 Jun 29. Plant Mol Biol. 2021. PMID: 34189708 Free PMC article.
-
The Absence of Hydrodynamic Stress Promotes Acquisition of Freezing Tolerance and Freeze-Dependent Asexual Reproduction in the Red Alga 'Bangia' sp. ESS1.Plants (Basel). 2021 Mar 1;10(3):465. doi: 10.3390/plants10030465. Plants (Basel). 2021. PMID: 33804533 Free PMC article.
References
-
- Baker N. R. (1991). A possible role for photosystem II in environmental perturbations of photosynthesis. Physiol. Plant 81, 563–570. 10.1111/j.1399-3054.1991.tb05101.x - DOI
-
- Bates L. S., Waldren R. P., Teare I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. 10.1007/BF00018060 - DOI
-
- Beike A. K., Lang D., Zimmer A. D., Wust F., Trautmann D., Wiedemann G., et al. . (2015). Insights from the cold transcriptome of Physcomitrella patens: global specialization pattern of conserved transcriptional regulators and identification of orphan genes involved in cold acclimation. New Phytol. 205, 869–881. 10.1111/nph.13004 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

