A new vector for efficient gene targeting to the pyrG locus in Aspergillus niger
- PMID: 28955454
- PMCID: PMC5611571
- DOI: 10.1186/s40694-015-0012-4
A new vector for efficient gene targeting to the pyrG locus in Aspergillus niger
Abstract
Background: The possibility for efficient gene targeting for the controlled integration of DNA constructs is an important tool in fungal genetics.
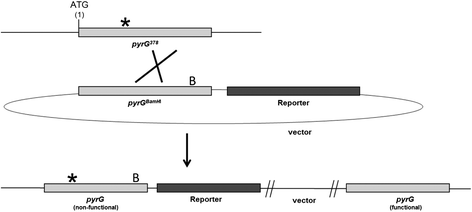
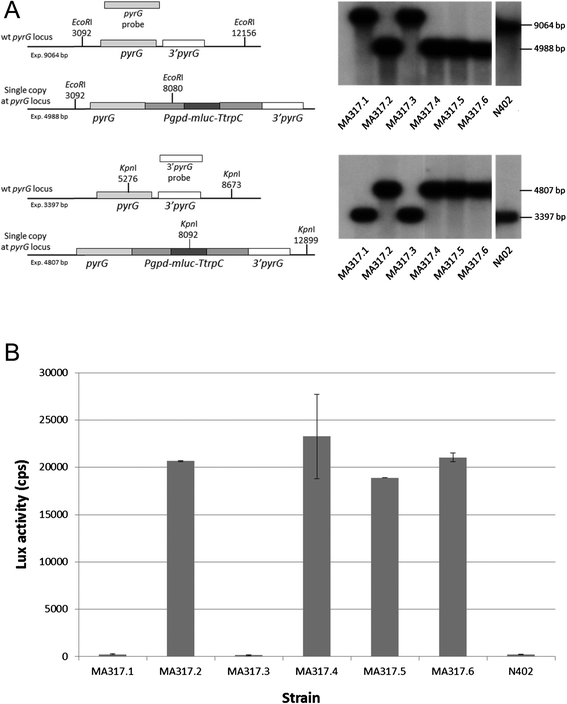
Findings: In this study, we report a new targeting vector based on the pyrG marker in Aspergillus niger. The DNA sequence to be targeted is surrounded by two fragments of the pyrG gene to allow homologous recombination of the recombinant DNA at the pyrG locus. The 5' end of the targeting cassette contains a non-functional truncated pyrG open reading frame (first 112 bases deleted) and the 3' untranslated region (3' UTR). At the 3' end, the targeting cassette consists of the 3' flanking region of the pyrG gene. A unique NotI site between the flanks allows the insertion of a gene of interest. The linearized targeting cassette is transformed to the A. niger pyrG mutant strain AB4.1 or a derivative thereof. By using a constitutively expressed luciferase reporter gene (mluc) as an example, it is shown that the targeting system is efficient as 4 out of 6 (67%) AB4.1 transformants and 51 out of 66 (77%) MA169.4 (ku70- ) transformants contained the reporter gene at the pyrG locus. A luciferase (lux) activity assay, performed with independently obtained transformants in which the mluc reporter was integrated at the pyrG locus, showed comparable and reproducible lux activities.
Conclusion: The new pyrG targeting vector is an important improvement to the existing method for gene targeting in A. niger. Although the vector is specific for A. niger, the presented design and approach is easily applicable for constructing integration vectors for other fungi.
Keywords: Luciferase activity; Promoter analysis; Reporter gene.
Figures

Similar articles
-
HisB as novel selection marker for gene targeting approaches in Aspergillus niger.BMC Microbiol. 2017 Mar 8;17(1):57. doi: 10.1186/s12866-017-0960-3. BMC Microbiol. 2017. PMID: 28274204 Free PMC article.
-
Development of a homologous transformation system for Aspergillus niger based on the pyrG gene.Mol Gen Genet. 1987 Jan;206(1):71-5. doi: 10.1007/BF00326538. Mol Gen Genet. 1987. PMID: 3472035
-
A novel strategy for the isolation of defined pyrG mutants and the development of a site-specific integration system for Aspergillus awamori.Curr Genet. 1995 May;27(6):536-40. doi: 10.1007/BF00314444. Curr Genet. 1995. PMID: 7553938
-
A gene transfer system based on the homologous pyrG gene and efficient expression of bacterial genes in Aspergillus oryzae.Curr Genet. 1989 Sep;16(3):159-63. doi: 10.1007/BF00391472. Curr Genet. 1989. PMID: 2688930
-
Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5'-decarboxylase gene, pyrG, as a unique transformation marker.Curr Genet. 1996 Jun;30(1):76-82. doi: 10.1007/s002940050103. Curr Genet. 1996. PMID: 8662213
Cited by
-
NHEJ and HDR can occur simultaneously during gene integration into the genome of Aspergillus niger.Fungal Biol Biotechnol. 2024 Aug 5;11(1):10. doi: 10.1186/s40694-024-00180-7. Fungal Biol Biotechnol. 2024. PMID: 39103967 Free PMC article.
-
HisB as novel selection marker for gene targeting approaches in Aspergillus niger.BMC Microbiol. 2017 Mar 8;17(1):57. doi: 10.1186/s12866-017-0960-3. BMC Microbiol. 2017. PMID: 28274204 Free PMC article.
-
Metabolomic Analysis of Aspergillus niger Isolated From the International Space Station Reveals Enhanced Production Levels of the Antioxidant Pyranonigrin A.Front Microbiol. 2020 May 21;11:931. doi: 10.3389/fmicb.2020.00931. eCollection 2020. Front Microbiol. 2020. PMID: 32670208 Free PMC article.
-
Construction of gene modification system with highly efficient and markerless for Monascus ruber M7.Front Microbiol. 2022 Aug 1;13:952323. doi: 10.3389/fmicb.2022.952323. eCollection 2022. Front Microbiol. 2022. PMID: 35979480 Free PMC article.
-
A set of isogenic auxotrophic strains for constructing multiple gene deletion mutants and parasexual crossings in Aspergillus niger.Arch Microbiol. 2016 Nov;198(9):861-8. doi: 10.1007/s00203-016-1240-6. Epub 2016 Jun 1. Arch Microbiol. 2016. PMID: 27251039 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

