Efficient gene editing in Neurospora crassa with CRISPR technology
- PMID: 28955455
- PMCID: PMC5611662
- DOI: 10.1186/s40694-015-0015-1
Efficient gene editing in Neurospora crassa with CRISPR technology
Abstract
Background: Efficient gene editing is a critical tool for investigating molecular mechanisms of cellular processes and engineering organisms for numerous purposes ranging from biotechnology to medicine. Recently developed RNA-guided CRISPR/Cas9 technology has been used for efficient gene editing in various organisms, but has not been tested in a model filamentous fungus, Neurospora crassa.
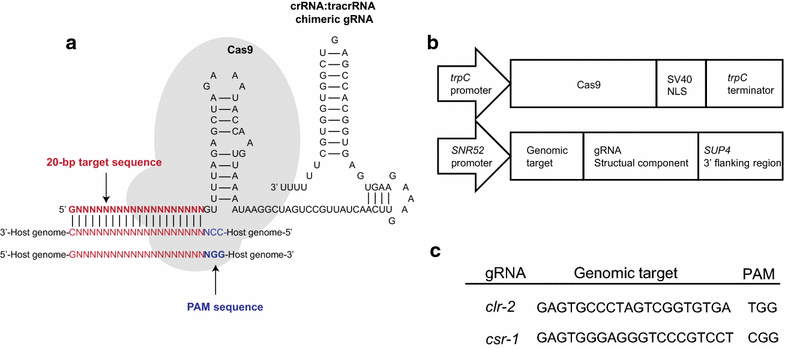
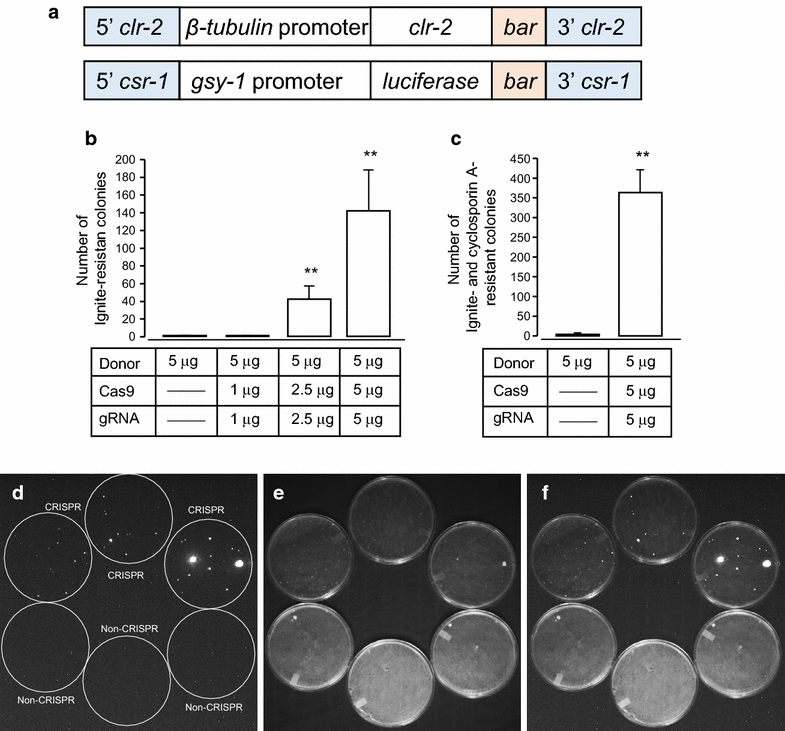
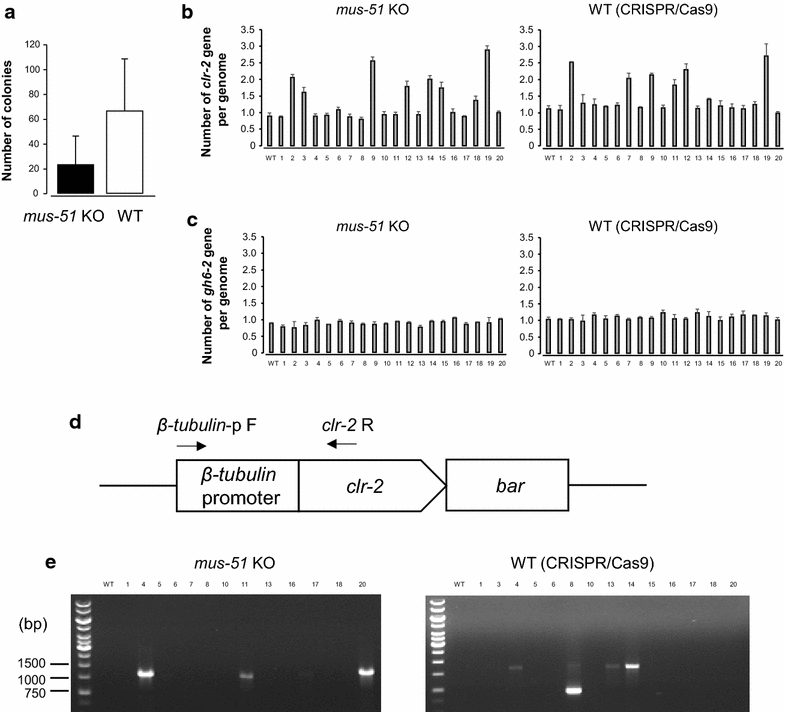
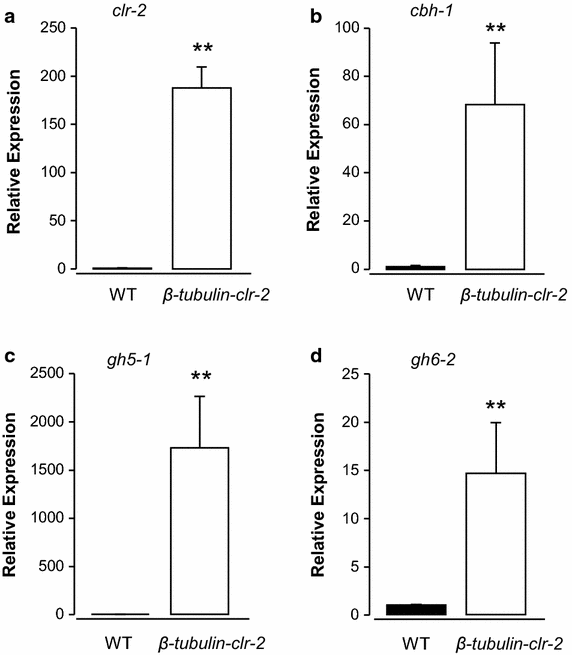
Findings: In this report, we demonstrate efficient gene replacement in a model filamentous fungus, Neurospora crassa, with the CRISPR/Cas9 system. We utilize Cas9 endonuclease and single crRNA:tracrRNA chimeric guide RNA (gRNA) to: (1) replace the endogenous promoter of clr-2 with the β-tubulin promoter, and (2) introduce a codon optimized fire fly luciferase under the control of the gsy-1 promoter at the csr-1 locus. CLR-2 is one of the core transcription factors that regulate the expression of cellulases, and GSY-1 regulates the conversion of glucose into glycogen. We show that the β-tubulin promoter driven clr-2 strain shows increased expression of cellulases, and gsy-1-luciferase reporter strain can be easily screened with a bioluminescence assay.
Conclusion: CRISPR/Cas9 system works efficiently in Neurospora crassa, which may be adapted to Neurospora natural isolates and other filamentous fungi. It will be beneficial for the filamentous fungal research community to take advantage of CRISPR/Cas9 tool kits that enable genetic perturbations including gene replacement and insertions.
Keywords: Biofuel; CRISPR/Cas9; Cellulase; Genome editing; Homologous recombination; Neurospora crassa.
Figures
Similar articles
-
Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system.Cell Discov. 2015 May 12;1:15007. doi: 10.1038/celldisc.2015.7. eCollection 2015. Cell Discov. 2015. PMID: 27462408 Free PMC article.
-
The transcriptional factor Clr-5 is involved in cellulose degradation through regulation of amino acid metabolism in Neurospora crassa.BMC Biotechnol. 2023 Nov 29;23(1):50. doi: 10.1186/s12896-023-00823-4. BMC Biotechnol. 2023. PMID: 38031036 Free PMC article.
-
Development of an Efficient C-to-T Base-Editing System and Its Application to Cellulase Transcription Factor Precise Engineering in Thermophilic Fungus Myceliophthora thermophila.Microbiol Spectr. 2022 Jun 29;10(3):e0232121. doi: 10.1128/spectrum.02321-21. Epub 2022 May 24. Microbiol Spectr. 2022. PMID: 35608343 Free PMC article.
-
Application of CRISPR in Filamentous Fungi and Macrofungi: From Component Function to Development Potentiality.ACS Synth Biol. 2023 Jul 21;12(7):1908-1923. doi: 10.1021/acssynbio.3c00099. Epub 2023 Jul 5. ACS Synth Biol. 2023. PMID: 37404005 Review.
-
CRISPR/Cas9 genome editing technology in filamentous fungi: progress and perspective.Appl Microbiol Biotechnol. 2019 Sep;103(17):6919-6932. doi: 10.1007/s00253-019-10007-w. Epub 2019 Jul 22. Appl Microbiol Biotechnol. 2019. PMID: 31332488 Free PMC article. Review.
Cited by
-
Cpf1 enables fast and efficient genome editing in Aspergilli.Fungal Biol Biotechnol. 2019 May 1;6:6. doi: 10.1186/s40694-019-0069-6. eCollection 2019. Fungal Biol Biotechnol. 2019. PMID: 31061713 Free PMC article.
-
Progress and Challenges: Development and Implementation of CRISPR/Cas9 Technology in Filamentous Fungi.Comput Struct Biotechnol J. 2019 Jun 13;17:761-769. doi: 10.1016/j.csbj.2019.06.007. eCollection 2019. Comput Struct Biotechnol J. 2019. PMID: 31312414 Free PMC article. Review.
-
Efficient genome editing in Fusarium oxysporum based on CRISPR/Cas9 ribonucleoprotein complexes.Fungal Genet Biol. 2018 Aug;117:21-29. doi: 10.1016/j.fgb.2018.05.003. Epub 2018 May 12. Fungal Genet Biol. 2018. PMID: 29763675 Free PMC article.
-
The freedom of choice.Fungal Biol Biotechnol. 2016 Oct 14;3:9. doi: 10.1186/s40694-016-0027-5. eCollection 2016. Fungal Biol Biotechnol. 2016. PMID: 28955468 Free PMC article. No abstract available.
-
Industrial applications of fungal lipases: a review.Front Microbiol. 2023 Apr 28;14:1142536. doi: 10.3389/fmicb.2023.1142536. eCollection 2023. Front Microbiol. 2023. PMID: 37187537 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

