Proteolytic activity assayed by subcellular localization switching of a substrate
- PMID: 28955937
- PMCID: PMC5613695
- DOI: 10.1016/j.bbrep.2016.07.011
Proteolytic activity assayed by subcellular localization switching of a substrate
Abstract
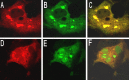
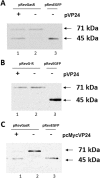
An approach to assay proteolytic activity in vivo by altering the subcellular localization of a labelled substrate was demonstrated. The assay included a protein shuttling between different cellular compartments and a site-specific recombinant protease. The shuttle protein used was the human immunodeficiency virus type 1 (HIV-1) Rev protein tandemly fused to the enhanced green fluorescent protein (EGFP) and the red fluorescent protein (RFP), while the protease was the site-specific protease VP24 from the herpes simplex virus type 1 (HSV-1). The fluorescent proteins in the Rev fusion protein were separated by a cleavage site specific for the VP24 protease. When co-expressed in COS-7 cells proteolysis was observed by fluorescence microscopy as a shift from a predominantly cytoplasmic localization of the fusion protein RevEGFP to a nuclear localization while the RFP part of the fusion protein remained in the cytoplasm. The cleavage of the fusion protein by VP24 was confirmed by Western blot analysis. The activity of VP24, when tagged N-terminally by the Myc-epitope, was found to be comparable to VP24. These results demonstrates that the activity and localization of a recombinantly expressed protease can be assessed by protease-mediated cleavage of fusion proteins containing a specific protease cleavage site.
Keywords: CLSM, confocal laser scanning microscopy; EGFP, enhanced green fluorescent protein; Green fluorescent protein; HIV, human immunodeficiency virus type 1; HIV-1 Rev; HSV-1 protease; HSV-1, herpes simplex virus type 1; HTLV-1, human T-cell leukaemia virus type 1; NES, nuclear export signal; NLS, nuclear localization signal; NOS, nucleolar localization signal; RFP, red fluorescent protein; Red fluorescent protein.
Figures

Similar articles
-
The Jembrana disease virus Rev protein: Identification of nuclear and novel lentiviral nucleolar localization and nuclear export signals.PLoS One. 2019 Aug 22;14(8):e0221505. doi: 10.1371/journal.pone.0221505. eCollection 2019. PLoS One. 2019. PMID: 31437223 Free PMC article.
-
The nuclear function of the nuclear diffusion inhibitory signal of human immunodeficiency virus type 1: critical roles in dominant nuclear localization and intracellular stability.J Hum Virol. 2000 Jul-Aug;3(4):173-81. J Hum Virol. 2000. PMID: 10990165
-
A set of enhanced green fluorescent protein concatemers for quantitative determination of nuclear localization signal strength.Anal Biochem. 2017 Sep 15;533:48-55. doi: 10.1016/j.ab.2017.06.015. Epub 2017 Jun 30. Anal Biochem. 2017. PMID: 28669708
-
Differential intracellular compartmentalization of herpetic thymidine kinases (TKs) in TK gene-transfected tumor cells: molecular characterization of the nuclear localization signal of herpes simplex virus type 1 TK.J Virol. 1998 Dec;72(12):9535-43. doi: 10.1128/JVI.72.12.9535-9543.1998. J Virol. 1998. PMID: 9811686 Free PMC article.
-
Restriction of human immunodeficiency virus type 1 Rev function in murine A9 cells involves the Rev C-terminal domain.J Virol. 2003 Mar;77(5):3084-90. doi: 10.1128/jvi.77.5.3084-3090.2003. J Virol. 2003. PMID: 12584334 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

