Effects of isoleucine 135 side chain length on the cofactor donor-acceptor distance within F420H2:NADP+ oxidoreductase: A kinetic analysis
- PMID: 28955995
- PMCID: PMC5614548
- DOI: 10.1016/j.bbrep.2016.11.012
Effects of isoleucine 135 side chain length on the cofactor donor-acceptor distance within F420H2:NADP+ oxidoreductase: A kinetic analysis
Abstract
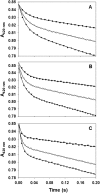
F420H2:NADP+ Oxidoreductase (Fno) catalyzes the reversible reduction of NADP+ to NADPH by transferring a hydride from the reduced F420 cofactor. Here, we have employed binding studies, steady-state and pre steady-state kinetic methods upon wtFno and isoleucine 135 (I135) Fno variants in order to study the effects of side chain length on the donor-acceptor distance between NADP+ and the F420 precursor, FO. The conserved I135 residue of Fno was converted to a valine, alanine and glycine, thereby shortening the side chain length. The steady-state kinetic analysis of wtFno and the variants showed classic Michaelis-Menten kinetics with varying FO concentrations. The data revealed a decreased kcat as side chain length decreased, with varying FO concentrations. The steady-state plots revealed non-Michaelis-Menten kinetic behavior when NADPH was varied. The double reciprocal plot of the varying NADPH concentrations displays a downward concave shape, while the NADPH binding curves gave Hill coefficients of less than 1. These data suggest that negative cooperativity occurs between the two identical monomers. The pre steady-state Abs420 versus time trace revealed biphasic kinetics, with a fast phase (hydride transfer) and a slow phase. The fast phase displayed an increased rate constant as side chain length decreased. The rate constant for the second phase, remained ~2 s-1 for each variant. Our data suggest that I135 plays a key role in sustaining the donor-acceptor distance between the two cofactors, thereby regulating the rate at which the hydride is transferred from FOH2 to NADP+. Therefore, Fno is a dynamic enzyme that regulates NADPH production.
Keywords: Dissociation constants; E. coli,, Escherichia coli; F420 cofactor; F420 cofactor, 7,8-didemethyl-8-hydroxy-5-deazariboflavin-5′-phosphoryllactyl(glutamyl)nglutamate, A. fulgidus, Archaeoglobus fulgidus; F420H2: NADP+ oxidoreductase; FO, precursor of F420 cofactor; Fno, F420H2:NADP+, oxidoreductase; Half-site reactivity; I135, Isoleucine 135; IPTG, isopropyl β-D-1-thiogalactopyranoside; Kd,, dissociation constant; Km, Michaelis-Menten constant; LB, Luria Bertani broth; NADP; NADP+, nicotinamide adenine dinucleotide phosphate; Negative cooperativity; PEI, Polyethyleneimine; Pre steady-state kinetics; Steady-state kinetics; k, rate constant; kcat, catalytic rate constant (turnover number), kcat /Km, catalytic efficiency; wtFno, wild-type Fno.
Figures





Similar articles
-
Evidence of Negative Cooperativity and Half-Site Reactivity within an F420-Dependent Enzyme: Kinetic Analysis of F420H2:NADP(+) Oxidoreductase.Biochemistry. 2016 Feb 23;55(7):1082-90. doi: 10.1021/acs.biochem.5b00762. Epub 2016 Feb 11. Biochemistry. 2016. PMID: 26811861
-
From Negative to No Cooperativity: Effects of Mutations on Intersubunit Communication within F420H2:NADP+ Oxidoreductase Using Steady-State and Pre-Steady-State Kinetic Methods.Biochemistry. 2025 Mar 18;64(6):1338-1347. doi: 10.1021/acs.biochem.4c00416. Epub 2025 Mar 4. Biochemistry. 2025. PMID: 40036042
-
Optimization of Expression and Purification of Recombinant Archeoglobus fulgidus F420H2:NADP+ Oxidoreductase, an F420 Cofactor Dependent Enzyme.Protein J. 2015 Dec;34(6):391-7. doi: 10.1007/s10930-015-9633-y. Protein J. 2015. PMID: 26493287
-
The multi-faceted role of NADPH in regulating inflammation in activated myeloid cells.Front Immunol. 2023 Dec 1;14:1328484. doi: 10.3389/fimmu.2023.1328484. eCollection 2023. Front Immunol. 2023. PMID: 38106413 Free PMC article. Review.
-
Regulatory Roles of Six-Transmembrane Epithelial Antigen of the Prostate Family Members in the Occurrence and Development of Malignant Tumors.Front Cell Dev Biol. 2021 Oct 29;9:752426. doi: 10.3389/fcell.2021.752426. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34778263 Free PMC article. Review.
Cited by
-
Structure/activity virtual screening and in vitro testing of small molecule inhibitors of 8-hydroxy-5-deazaflavin:NADPH oxidoreductase from gut methanogenic bacteria.Sci Rep. 2020 Aug 4;10(1):13150. doi: 10.1038/s41598-020-70042-w. Sci Rep. 2020. PMID: 32753591 Free PMC article.
References
-
- Kunow J., Schwörer B., Stetter K.O., Thauer R.K. A F420-dependent NADP reductase in the extremely thermophilic sulfate-reducing Archaeoglobus fulgidus. Arch. Microbiol. 1993;160:199–205.
-
- Berk H., Thauer R.K. F420H2:NADP oxidoreductase from Methanobacterium thermoautotrophicum: identification of the encoding gene via functional overexpression in Escherichia coli. FEBS Lett. 1998;438:124–126. - PubMed
-
- Yamazaki S., Tsai L. Purification and properties of 8-hydroxy-5-deazaflavin-dependent NADP+ reductase from Methanococcus vannielii. J. Biol. Chem. 1980;255:6462–6465. - PubMed
-
- Berk H., Thauer R.K. Function of coenzyme F420-dependent NADP reductase in methanogenic archaea containing an NADP-dependent alcohol dehydrogenase. Arch. Microbiol. 1997;168:396–402. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

