Quantitative and Systems Pharmacology. 1. In Silico Prediction of Drug-Target Interactions of Natural Products Enables New Targeted Cancer Therapy
- PMID: 28956927
- PMCID: PMC5971208
- DOI: 10.1021/acs.jcim.7b00216
Quantitative and Systems Pharmacology. 1. In Silico Prediction of Drug-Target Interactions of Natural Products Enables New Targeted Cancer Therapy
Abstract
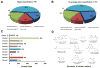
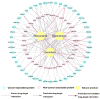
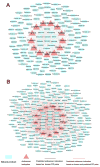
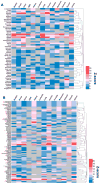
Natural products with diverse chemical scaffolds have been recognized as an invaluable source of compounds in drug discovery and development. However, systematic identification of drug targets for natural products at the human proteome level via various experimental assays is highly expensive and time-consuming. In this study, we proposed a systems pharmacology infrastructure to predict new drug targets and anticancer indications of natural products. Specifically, we reconstructed a global drug-target network with 7,314 interactions connecting 751 targets and 2,388 natural products and built predictive network models via a balanced substructure-drug-target network-based inference approach. A high area under receiver operating characteristic curve of 0.96 was yielded for predicting new targets of natural products during cross-validation. The newly predicted targets of natural products (e.g., resveratrol, genistein, and kaempferol) with high scores were validated by various literature studies. We further built the statistical network models for identification of new anticancer indications of natural products through integration of both experimentally validated and computationally predicted drug-target interactions of natural products with known cancer proteins. We showed that the significantly predicted anticancer indications of multiple natural products (e.g., naringenin, disulfiram, and metformin) with new mechanism-of-action were validated by various published experimental evidence. In summary, this study offers powerful computational systems pharmacology approaches and tools for the development of novel targeted cancer therapies by exploiting the polypharmacology of natural products.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
In silico polypharmacology of natural products.Brief Bioinform. 2018 Nov 27;19(6):1153-1171. doi: 10.1093/bib/bbx045. Brief Bioinform. 2018. PMID: 28460068 Review.
-
Quantitative and systems pharmacology 2. In silico polypharmacology of G protein-coupled receptor ligands via network-based approaches.Pharmacol Res. 2018 Mar;129:400-413. doi: 10.1016/j.phrs.2017.11.005. Epub 2017 Nov 10. Pharmacol Res. 2018. PMID: 29133212
-
Systems pharmacology strategies for anticancer drug discovery based on natural products.Mol Biosyst. 2014 Jul;10(7):1912-7. doi: 10.1039/c4mb00105b. Epub 2014 May 6. Mol Biosyst. 2014. PMID: 24802653
-
In silico prediction of chemical mechanism of action via an improved network-based inference method.Br J Pharmacol. 2016 Dec;173(23):3372-3385. doi: 10.1111/bph.13629. Epub 2016 Nov 1. Br J Pharmacol. 2016. PMID: 27646592 Free PMC article.
-
In Silico Oncology Drug Repositioning and Polypharmacology.Methods Mol Biol. 2019;1878:243-261. doi: 10.1007/978-1-4939-8868-6_15. Methods Mol Biol. 2019. PMID: 30378081 Review.
Cited by
-
Molecular dynamics simulations, docking and MMGBSA studies of newly designed peptide-conjugated glucosyloxy stilbene derivatives with tumor cell receptors.Mol Divers. 2022 Oct;26(5):2717-2743. doi: 10.1007/s11030-021-10354-9. Epub 2022 Jan 17. Mol Divers. 2022. PMID: 35037187
-
A Systems Pharmacology Approach Uncovers Wogonoside as an Angiogenesis Inhibitor of Triple-Negative Breast Cancer by Targeting Hedgehog Signaling.Cell Chem Biol. 2019 Aug 15;26(8):1143-1158.e6. doi: 10.1016/j.chembiol.2019.05.004. Epub 2019 Jun 6. Cell Chem Biol. 2019. PMID: 31178408 Free PMC article.
-
Closing the Gap Between Therapeutic Use and Mode of Action in Remedial Herbs.Front Pharmacol. 2019 Oct 3;10:1132. doi: 10.3389/fphar.2019.01132. eCollection 2019. Front Pharmacol. 2019. PMID: 31632273 Free PMC article.
-
In Silico Pharmacoepidemiologic Evaluation of Drug-Induced Cardiovascular Complications Using Combined Classifiers.J Chem Inf Model. 2018 May 29;58(5):943-956. doi: 10.1021/acs.jcim.7b00641. Epub 2018 May 10. J Chem Inf Model. 2018. PMID: 29712429 Free PMC article.
-
Systems pharmacology dissection of pharmacological mechanisms of Xiaochaihu decoction against human coronavirus.BMC Complement Med Ther. 2023 Jul 20;23(1):252. doi: 10.1186/s12906-023-04024-6. BMC Complement Med Ther. 2023. PMID: 37475019 Free PMC article.
References
-
- DeCorte BL. Underexplored Opportunities for Natural Products in Drug Discovery. J Med Chem. 2016;59:9295–9304. - PubMed
-
- Harvey AL, Edrada-Ebel R, Quinn RJ. The Re-emergence of Natural Products for Drug Discovery in the Genomics Era. Nat Rev Drug Discovery. 2015;14:111–129. - PubMed
-
- Li JW, Vederas JC. Drug Discovery and Natural Products: End of an Era or an Endless Frontier? Science. 2009;325:161–165. - PubMed
-
- Rodrigues T, Reker D, Schneider P, Schneider G. Counting on Natural Products for Drug Design. Nat Chem. 2016;8:531–541. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

