The diversity of the Chagas parasite, Trypanosoma cruzi, infecting the main Central American vector, Triatoma dimidiata, from Mexico to Colombia
- PMID: 28957315
- PMCID: PMC5619707
- DOI: 10.1371/journal.pntd.0005878
The diversity of the Chagas parasite, Trypanosoma cruzi, infecting the main Central American vector, Triatoma dimidiata, from Mexico to Colombia
Abstract
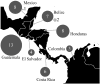
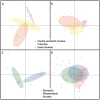
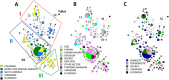
Little is known about the strains of Trypanosoma cruzi circulating in Central America and specifically in the most important vector in this region, Triatoma dimidiata. Approximately six million people are infected with T. cruzi, the causative agent of Chagas disease, which has the greatest negative economic impact and is responsible for ~12,000 deaths annually in Latin America. By international consensus, strains of T. cruzi are divided into six monophyletic clades called discrete typing units (DTUs TcI-VI) and a seventh DTU first identified in bats called TcBat. TcI shows the greatest geographic range and diversity. Identifying strains present and diversity within these strains is important as different strains and their genotypes may cause different pathologies and may circulate in different localities and transmission cycles, thus impacting control efforts, treatment and vaccine development. To determine parasite strains present in T. dimidiata across its geographic range from Mexico to Colombia, we isolated abdominal DNA from T. dimidiata and determined which specimens were infected with T. cruzi by PCR. Strains from infected insects were determined by comparing the sequence of the 18S rDNA and the spliced-leader intergenic region to typed strains in GenBank. Two DTUs were found: 94% of infected T. dimidiata contained TcI and 6% contained TcIV. TcI exhibited high genetic diversity. Geographic structure of TcI haplotypes was evident by Principal Component and Median-Joining Network analyses as well as a significant result in the Mantel test, indicating isolation by distance. There was little evidence of association with TcI haplotypes and host/vector or ecotope. This study provides new information about the strains circulating in the most important Chagas vector in Central America and reveals considerable variability within TcI as well as geographic structuring at this large geographic scale. The lack of association with particular vectors/hosts or ecotopes suggests the parasites are moving among vectors/hosts and ecotopes therefore a comprehensive approach, such as the Ecohealth approach that makes houses refractory to the vectors will be needed to successfully halt transmission of Chagas disease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Molecular Diversity of Trypanosoma cruzi Detected in the Vector Triatoma protracta from California, USA.PLoS Negl Trop Dis. 2016 Jan 21;10(1):e0004291. doi: 10.1371/journal.pntd.0004291. eCollection 2016 Jan. PLoS Negl Trop Dis. 2016. PMID: 26797311 Free PMC article.
-
Untangling the transmission dynamics of primary and secondary vectors of Trypanosoma cruzi in Colombia: parasite infection, feeding sources and discrete typing units.Parasit Vectors. 2016 Dec 1;9(1):620. doi: 10.1186/s13071-016-1907-5. Parasit Vectors. 2016. PMID: 27903288 Free PMC article.
-
The Chagas disease domestic transmission cycle in Guatemala: Parasite-vector switches and lack of mitochondrial co-diversification between Triatoma dimidiata and Trypanosoma cruzi subpopulations suggest non-vectorial parasite dispersal across the Motagua valley.Acta Trop. 2015 Nov;151:80-7. doi: 10.1016/j.actatropica.2015.07.014. Epub 2015 Jul 26. Acta Trop. 2015. PMID: 26215126
-
Triatoma dimidiata (Latreille, 1811): a review of its diversity across its geographic range and the relationship among populations.Infect Genet Evol. 2007 Mar;7(2):343-52. doi: 10.1016/j.meegid.2006.10.001. Epub 2006 Nov 13. Infect Genet Evol. 2007. PMID: 17097928 Review.
-
Trypanosoma cruzi genetic diversity: Something new for something known about Chagas disease manifestations, serodiagnosis and drug sensitivity.Acta Trop. 2018 Aug;184:38-52. doi: 10.1016/j.actatropica.2017.09.017. Epub 2017 Sep 21. Acta Trop. 2018. PMID: 28941731 Review.
Cited by
-
Molecular Markers for the Phylogenetic Reconstruction of Trypanosoma cruzi: A Quantitative Review.Pathogens. 2025 Jan 14;14(1):72. doi: 10.3390/pathogens14010072. Pathogens. 2025. PMID: 39861033 Free PMC article. Review.
-
Motility-based label-free detection of parasites in bodily fluids using holographic speckle analysis and deep learning.Light Sci Appl. 2018 Dec 12;7:108. doi: 10.1038/s41377-018-0110-1. eCollection 2018. Light Sci Appl. 2018. PMID: 30564314 Free PMC article.
-
Spatial metabolomics identifies localized chemical changes in heart tissue during chronic cardiac Chagas Disease.PLoS Negl Trop Dis. 2021 Oct 4;15(10):e0009819. doi: 10.1371/journal.pntd.0009819. eCollection 2021 Oct. PLoS Negl Trop Dis. 2021. PMID: 34606502 Free PMC article.
-
Animal Models of Trypanosoma cruzi Congenital Transmission.Pathogens. 2022 Oct 11;11(10):1172. doi: 10.3390/pathogens11101172. Pathogens. 2022. PMID: 36297229 Free PMC article. Review.
-
Biological and Molecular Characterization of Five Trypanosoma cruzi (Chagas, 1909) (Kinetoplastida, Trypanosomatidae) Isolates from the State of Hidalgo, Mexico.Trop Med Infect Dis. 2025 May 1;10(5):122. doi: 10.3390/tropicalmed10050122. Trop Med Infect Dis. 2025. PMID: 40423352 Free PMC article.
References
-
- World Health Organization. Weekly Epidemiology Report: Chagas disease in Latin America: an epidemiological update based on 2010 estimates. Switzerland: 2015. - PubMed
-
- Tibayrenc M. Genetic subdivisions within Trypanosoma cruzi (Discrete Typing Units) and their relevance for molecular epidemiology and experimental evolution. Kinetoplastid biology and disease. 2003;2(1):12 doi: 10.1186/1475-9292-2-12 - DOI - PMC - PubMed
-
- Messenger LA, Miles MA. Evidence and importance of genetic exchange among field populations of Trypanosoma cruzi. Acta Trop. 2015;151:150–5. doi: 10.1016/j.actatropica.2015.05.007 - DOI - PMC - PubMed
-
- Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MM, et al. The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol. 2012;12(2):240–53. doi: 10.1016/j.meegid.2011.12.009 . - DOI - PubMed
-
- Marcili A, Lima L, Cavazzana M, Junqueira AC, Veludo HH, Maia Da Silva F, et al. A new genotype of Trypanosoma cruzi associated with bats evidenced by phylogenetic analyses using SSU rDNA, cytochrome b and Histone H2B genes and genotyping based on ITS1 rDNA. Parasitology. 2009;136(6):641–55. doi: 10.1017/S0031182009005861 . - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

