Interactions between the tumor and the blood systemic response of breast cancer patients
- PMID: 28957325
- PMCID: PMC5619688
- DOI: 10.1371/journal.pcbi.1005680
Interactions between the tumor and the blood systemic response of breast cancer patients
Abstract
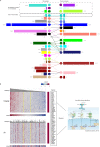
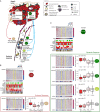
Although systemic immunity is critical to the process of tumor rejection, cancer research has largely focused on immune cells in the tumor microenvironment. To understand molecular changes in the patient systemic response (SR) to the presence of BC, we profiled RNA in blood and matched tumor from 173 patients. We designed a system (MIxT, Matched Interactions Across Tissues) to systematically explore and link molecular processes expressed in each tissue. MIxT confirmed that processes active in the patient SR are especially relevant to BC immunogenicity. The nature of interactions across tissues (i.e. which biological processes are associated and their patterns of expression) varies highly with tumor subtype. For example, aspects of the immune SR are underexpressed proportionally to the level of expression of defined molecular processes specific to basal tumors. The catalog of subtype-specific interactions across tissues from BC patients provides promising new ways to tackle or monitor the disease by exploiting the patient SR.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Yarden Y. The biological framework: translational research from bench to clinic. Oncologist. 2010;15 Suppl 5:1–7. doi: 10.1634/theoncologist.2010-S5-01 . - DOI - PubMed
-
- Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol. 2010;220(2):263–80. Epub 2009/11/21. doi: 10.1002/path.2648 . - DOI - PubMed
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. Epub 2000/08/30. doi: 10.1038/35021093 . - DOI - PubMed
-
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(19):10869–74. Epub 2001/09/13. doi: 10.1073/pnas.191367098 ; PubMed Central PMCID: PMC58566. - DOI - PMC - PubMed
-
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27(8):1160–7. Epub 2009/02/11. doi: 10.1200/JCO.2008.18.1370 ; PubMed Central PMCID: PMC2667820. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

