Gene Regulatory Network Inference from Single-Cell Data Using Multivariate Information Measures
- PMID: 28957658
- PMCID: PMC5624513
- DOI: 10.1016/j.cels.2017.08.014
Gene Regulatory Network Inference from Single-Cell Data Using Multivariate Information Measures
Abstract
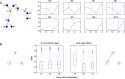
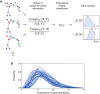
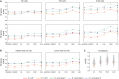
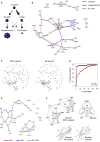
While single-cell gene expression experiments present new challenges for data processing, the cell-to-cell variability observed also reveals statistical relationships that can be used by information theory. Here, we use multivariate information theory to explore the statistical dependencies between triplets of genes in single-cell gene expression datasets. We develop PIDC, a fast, efficient algorithm that uses partial information decomposition (PID) to identify regulatory relationships between genes. We thoroughly evaluate the performance of our algorithm and demonstrate that the higher-order information captured by PIDC allows it to outperform pairwise mutual information-based algorithms when recovering true relationships present in simulated data. We also infer gene regulatory networks from three experimental single-cell datasets and illustrate how network context, choices made during analysis, and sources of variability affect network inference. PIDC tutorials and open-source software for estimating PID are available. PIDC should facilitate the identification of putative functional relationships and mechanistic hypotheses from single-cell transcriptomic data.
Keywords: gene regulation; mutual information; network reconstruction; single-cell PCR; single-cell RNA-seq.
Copyright © 2017. Published by Elsevier Inc.
Figures


Similar articles
-
Gene Regulatory Networks from Single Cell Data for Exploring Cell Fate Decisions.Methods Mol Biol. 2019;1975:211-238. doi: 10.1007/978-1-4939-9224-9_10. Methods Mol Biol. 2019. PMID: 31062312
-
Inferring Causal Gene Regulatory Networks from Coupled Single-Cell Expression Dynamics Using Scribe.Cell Syst. 2020 Mar 25;10(3):265-274.e11. doi: 10.1016/j.cels.2020.02.003. Epub 2020 Mar 4. Cell Syst. 2020. PMID: 32135093 Free PMC article.
-
Evaluating methods of inferring gene regulatory networks highlights their lack of performance for single cell gene expression data.BMC Bioinformatics. 2018 Jun 19;19(1):232. doi: 10.1186/s12859-018-2217-z. BMC Bioinformatics. 2018. PMID: 29914350 Free PMC article.
-
Learning Differential Module Networks Across Multiple Experimental Conditions.Methods Mol Biol. 2019;1883:303-321. doi: 10.1007/978-1-4939-8882-2_13. Methods Mol Biol. 2019. PMID: 30547406 Review.
-
A comprehensive survey of regulatory network inference methods using single cell RNA sequencing data.Brief Bioinform. 2021 May 20;22(3):bbaa190. doi: 10.1093/bib/bbaa190. Brief Bioinform. 2021. PMID: 34020546 Free PMC article. Review.
Cited by
-
Single cell metabolism: current and future trends.Metabolomics. 2022 Oct 1;18(10):77. doi: 10.1007/s11306-022-01934-3. Metabolomics. 2022. PMID: 36181583 Free PMC article. Review.
-
Approximate Bayesian computation for inferring Waddington landscapes from single-cell data.R Soc Open Sci. 2024 Jul 10;11(7):231697. doi: 10.1098/rsos.231697. eCollection 2024 Jul. R Soc Open Sci. 2024. PMID: 39076359 Free PMC article.
-
Holimap: an accurate and efficient method for solving stochastic gene network dynamics.Nat Commun. 2024 Aug 2;15(1):6557. doi: 10.1038/s41467-024-50716-z. Nat Commun. 2024. PMID: 39095346 Free PMC article.
-
In vivo single-cell lineage tracing in zebrafish using high-resolution infrared laser-mediated gene induction microscopy.Elife. 2020 Jan 6;9:e52024. doi: 10.7554/eLife.52024. Elife. 2020. PMID: 31904340 Free PMC article.
-
Unraveling T Cell Responses for Long Term Protection of SARS-CoV-2 Infection.Front Genet. 2022 May 4;13:871164. doi: 10.3389/fgene.2022.871164. eCollection 2022. Front Genet. 2022. PMID: 35601483 Free PMC article.
References
-
- Agresti A., Hitchcock D.B. Bayesian inference for categorical data analysis. Stat. Methods Appt. 2005;14:297–330.
-
- Beal M.J., Falciani F., Ghahramani Z., Rangel C., Wild D.L. A Bayesian approach to reconstructing genetic regulatory networks with hidden factors. Bioinformatics. 2005;21:349–356. - PubMed
-
- Bezanson J., Edelman A., Karpinski S., Shah V.B. Julia: a fresh approach to numerical computing. arXiv. 2014 1411.1607.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

