Stable Isotope Labeling Reveals Novel Insights Into Ubiquitin-Mediated Protein Aggregation With Age, Calorie Restriction, and Rapamycin Treatment
- PMID: 28958078
- PMCID: PMC6380815
- DOI: 10.1093/gerona/glx047
Stable Isotope Labeling Reveals Novel Insights Into Ubiquitin-Mediated Protein Aggregation With Age, Calorie Restriction, and Rapamycin Treatment
Abstract
Accumulation of protein aggregates with age was first described in aged human tissue over 150 years ago and has since been described in virtually every human tissue. Ubiquitin modifications are a canonical marker of insoluble protein aggregates; however, the composition of most age-related inclusions remains relatively unknown. To examine the landscape of age-related protein aggregation in vivo, we performed an antibody-based pulldown of ubiquitinated proteins coupled with metabolic labeling and mass spectrometry on young and old mice on calorie restriction (CR), rapamycin (RP)-supplemented, and control diets. We show increased abundance of many ubiquitinated proteins in old mice and greater retention of preexisting (unlabeled) ubiquitinated proteins relative to their unmodified counterparts-fitting the expected profile of age-increased accumulation of long-lived aggregating proteins. Both CR and RP profoundly affected ubiquitinome composition, half-live, and the insolubility of proteins, consistent with their ability to mobilize these age-associated accumulations. Finally, confocal microscopy confirmed the aggregation of two of the top predicted aggregating proteins, keratins 8/18 and catalase, as well as their attenuation by CR and RP. Stable-isotope labeling is a powerful tool to gain novel insights into proteostasis mechanisms, including protein aggregation, and could be used to identify novel therapeutic targets in aging and protein aggregation diseases.
Figures



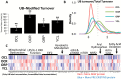

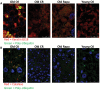
Comment in
-
Identifying ubiquitinated proteins and aggregates.Aging (Albany NY). 2018 Oct 18;10(10):2549-2550. doi: 10.18632/aging.101605. Aging (Albany NY). 2018. PMID: 30335616 Free PMC article. No abstract available.
References
-
- Hedhli N, Pelat M, Depre C. Protein turnover in cardiac cell growth and survival. Cardiovasc Res. 2005;68:186–196. doi:10.1016/j.cardiores.2005.06.025 - PubMed
-
- Surguchev A, Surguchov A. Conformational diseases: looking into the eyes. Brain Res Bull. 2010;81:12–24. doi:10.1016/j.brainresbull.2009.09.015 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

