Deciphering of the Human Interferon-Regulated Proteome by Mass Spectrometry-Based Quantitative Analysis Reveals Extent and Dynamics of Protein Induction and Repression
- PMID: 28959263
- PMCID: PMC5603615
- DOI: 10.3389/fimmu.2017.01139
Deciphering of the Human Interferon-Regulated Proteome by Mass Spectrometry-Based Quantitative Analysis Reveals Extent and Dynamics of Protein Induction and Repression
Abstract
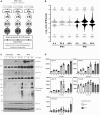
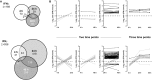
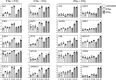
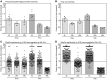
Interferons (IFNs) are pleotropic cytokines secreted upon encounter of pathogens and tumors. Applying their antipathogenic, antiproliferative, and immune stimulatory capacities, recombinant IFNs are frequently prescribed as drugs to treat different diseases. IFNs act by changing the gene expression profile of cells. Due to characteristics such as rapid gene induction and signaling, IFNs also represent prototypical model systems for various aspects of biomedical research (e.g., signal transduction). In regard to the signaling and activated promoters, IFNs can be subdivided into two groups. Here, alterations of the cellular proteome of human cells treated with IFNα and IFNγ were elucidated in a time-resolved manner by quantitative proteome analysis. The majority of protein regulations were strongly IFN type and time dependent. In addition to the expected upregulation of IFN-responsive proteins, an astonishing number of proteins became profoundly repressed especially by IFNγ. Thus, our comprehensive analysis revealed important insights into the human IFN-regulated proteome and its dynamics of protein induction and repression. Interestingly, the new class of IFN-repressed genes comprises known host factors for highly relevant pathogens such as HIV, dengue virus, and hepatitis C virus.
Keywords: IFN-repressed gene; IFN-stimulated gene; IFNalpha; IFNgamma; interferon; mass spectrometry; proteome.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

