How do lncRNAs regulate transcription?
- PMID: 28959731
- PMCID: PMC5617379
- DOI: 10.1126/sciadv.aao2110
How do lncRNAs regulate transcription?
Abstract
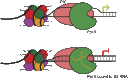
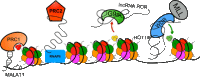
It has recently become apparent that RNA, itself the product of transcription, is a major regulator of the transcriptional process. In particular, long noncoding RNAs (lncRNAs), which are so numerous in eukaryotes, function in many cases as transcriptional regulators. These RNAs function through binding to histone-modifying complexes, to DNA binding proteins (including transcription factors), and even to RNA polymerase II. In other cases, it is the act of lncRNA transcription rather than the lncRNA product that appears to be regulatory. We review recent progress in elucidating the molecular mechanisms by which lncRNAs modulate gene expression and future opportunities in this research field.
Figures
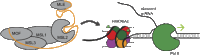

References
-
- Liu S. J., Horlbeck M. A., Cho S. W., Birk H. S., Malatesta M., He D., Attenello F. J., Villalta J. E., Cho M. Y., Chen Y., Mandegar M. A., Olvera M. P., Gilbert L. A., Conklin B. R., Chang H. Y., Weissman J. S., Lim D. A., CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, eaah7111 (2017). - PMC - PubMed
-
- Li W., Notani D., Rosenfeld M. G., Enhancers as non-coding RNA transcription units: Recent insights and future perspectives. Nat. Rev. Genet. 17, 207–223 (2016). - PubMed
-
- Cech T. R., Steitz J. A., The noncoding RNA revolution—Trashing old rules to forge new ones. Cell 157, 77–94 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

