Deubiquitylating enzymes and drug discovery: emerging opportunities
- PMID: 28959952
- PMCID: PMC7097658
- DOI: 10.1038/nrd.2017.152
Deubiquitylating enzymes and drug discovery: emerging opportunities
Abstract
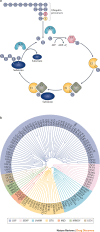
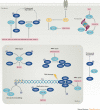
More than a decade after a Nobel Prize was awarded for the discovery of the ubiquitin-proteasome system and clinical approval of proteasome and ubiquitin E3 ligase inhibitors, first-generation deubiquitylating enzyme (DUB) inhibitors are now approaching clinical trials. However, although our knowledge of the physiological and pathophysiological roles of DUBs has evolved tremendously, the clinical development of selective DUB inhibitors has been challenging. In this Review, we discuss these issues and highlight recent advances in our understanding of DUB enzymology and biology as well as technological improvements that have contributed to the current interest in DUBs as therapeutic targets in diseases ranging from oncology to neurodegeneration.
Conflict of interest statement
S.P.J. is part-time employed by Mission Therapeutics, Cambridge, UK, owns shares and is a director of the company. J.A.H. is a full-time employee of Mission Therapeutics. N.M.M. and X.J. own shares in Mission Therapeutics.
Figures
References
-
- Ciechanover A. The ubiquitin proteolytic system and pathogenesis of human diseases: a novel platform for mechanism-based drug targeting. Biochem. Soc. Trans. 2003;31:474–481. - PubMed
-
- Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 2005;6:79–87. - PubMed
-
- Gallastegui N, Groll M. The 26S proteasome: assembly and function of a destructive machine. Trends Biochem. Sci. 2010;35:634–642. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

