Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN
- PMID: 28959974
- PMCID: PMC5624508
- DOI: 10.1038/nature24023
Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN
Abstract
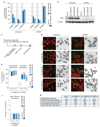
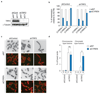
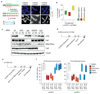
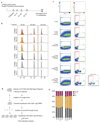
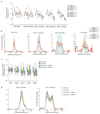
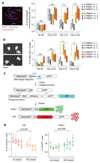
Classical non-homologous end joining (cNHEJ) and homologous recombination compete for the repair of double-stranded DNA breaks during the cell cycle. Homologous recombination is inhibited during the G1 phase of the cell cycle, but both pathways are active in the S and G2 phases. However, it is unclear why cNHEJ does not always outcompete homologous recombination during the S and G2 phases. Here we show that CYREN (cell cycle regulator of NHEJ) is a cell-cycle-specific inhibitor of cNHEJ. Suppression of CYREN allows cNHEJ to occur at telomeres and intrachromosomal breaks during the S and G2 phases, and cells lacking CYREN accumulate chromosomal aberrations upon damage induction, specifically outside the G1 phase. CYREN acts by binding to the Ku70/80 heterodimer and preferentially inhibits cNHEJ at breaks with overhangs by protecting them. We therefore propose that CYREN is a direct cell-cycle-dependent inhibitor of cNHEJ that promotes error-free repair by homologous recombination during cell cycle phases when sister chromatids are present.
Conflict of interest statement
Author Information
The authors declare no competing financial interests.
Figures



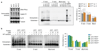




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

